Difference between revisions of "Welcome to the In-Silico Model of Cutaneous Lipids Wiki"
(→Reactions) |
(→Description of the Model) |
||
| (38 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| − | + | == Description of the Model == | |
| + | Computational models have been demonstrated as useful tools to gain a better understanding of networks, aid rational decision-making and support research across the biosciences<ref>Yang, K. Ma, W. Liang, H. Ouyang, Q. Tang, C. Lai, L., ''Dynamic simulations on the arachidonic acid metabolic network'', PLoS Comput Biol (2007), 3, e55</ref><ref>Gupta, S. Maurya, M. R. Stephens, D. L. Dennis, E. A. Subramaniam, S., ''An integrated model of eicosanoid metabolism and signalling based on lipidomics flux analysis'', Biophys J (2009), 96, 4542-51.</ref><ref>Dobovisek, A. Fajmut, A. Brumen, M., ''Role of expression of prostaglandin synthases 1 and 2 and leukotriene C4 synthase in aspirin-intolerant asthma: a theoretical study'', J Pharmacokinet Pharmacodyn (2011), 38, 261-78.</ref>. Due to the complexity of the AA cascade, predicting the outcome of the network is difficult due to the collaborative action of numerous species (i.e. metabolites, proteins, signalling cascades). In addition, the AA cascade is ubiquitous and is implicated in numerous inflammatory diseases such as cancer, atherosclerosis and asthma <ref>Bishop-Bailey, D. Calatayud, S. Warner, T. D. Hla, T. Mitchell, J. A., ''Prostaglandins and the regulation of tumor growth'', J Environ Pathol Toxicol Oncol (2002), 21, 93-101</ref><ref>von Schacky, C., ''Omega-3 fatty acids in cardiovascular disease - an uphill battle.'', Prostaglandins Leukot Essent Fatty Acids (2015), 92, 41-7.</ref> <ref>Claar, D. Hartert, T. V. Peebles, R. S., Jr., ''The role of prostaglandins in allergic lung inflammation and asthma'', Expert Rev Respir Med (2015), 9, 55-72.</ref><ref>Albanesi, C. Pastore, S., ''Pathobiology of chronic inflammatory skin diseases: interplay between keratinocytes and immune cells as a target for anti-inflammatory drugs'', Curr Drug Metab (2010), 11, 210-27.</ref>; as a result, the production of eicosanoids is the targets of several pharmacological interventions.Therefore, there is a need to utilise a systems-level approach to further investigate the bioactive AA cascade. This can be achieved by building upon previous models of the AA cascade <ref>Yang, K. Ma, W. Liang, H. Ouyang, Q. Tang, C. Lai, L., ''Dynamic simulations on the arachidonic acid metabolic network'', PLoS Comput Biol (2007), 3, e55</ref><ref>Gupta, S. Maurya, M. R. Stephens, D. L. Dennis, E. A. Subramaniam, S., ''An integrated model of eicosanoid metabolism and signalling based on lipidomics flux analysis'', Biophys J (2009), 96, 4542-51.</ref><ref>Dobovisek, A. Fajmut, A. Brumen, M., ''Role of expression of prostaglandin synthases 1 and 2 and leukotriene C4 synthase in aspirin-intolerant asthma: a theoretical study'', J Pharmacokinet Pharmacodyn (2011), 38, 261-78.</ref> and creating a new model which addresses the gaps in the literature such as an extensive network topology, the use of experimental parameters, documentation of parameter sources, integration of data from multiple platforms and application beyond a single cell type. | ||
| − | + | Furthermore, there is a need to incorporate novel approaches to the parameterisation and simulation of metabolic models, in order to quantify the uncertainty associated with literature data and indicate the robustness of each prediction <ref>Tsigkinopoulou, A. Hawari, A. Uttley, M. Breitling, R., ''Defining informative priors for ensemble modeling in systems biology'', Nat Protoc (2018), 13, 2643-2663.</ref>. The main aim of this project is to construct, analyse and validate a predictive and adaptable metabolic model of the AA cascade. To use this model to investigate how the concentration of metabolites change over time following internal/external perturbations, we need to utilise a kinetic deterministic modelling approach. This work will build upon the previous models of the AA cascade and support researchers across the biosciences to gain a better understanding of pathways by incorporating means of adapting the model to represent different cell types. We will demonstrate the application of this work in immortalised human cell lines, HaCaT keratinocytes and 46BR.1N fibroblasts as they are shown to produce different eicosanoids, availabilities of AA, protein profiles and response to inflammatory stimuli. | |
| − | |||
| − | |||
| − | |||
| − | |||
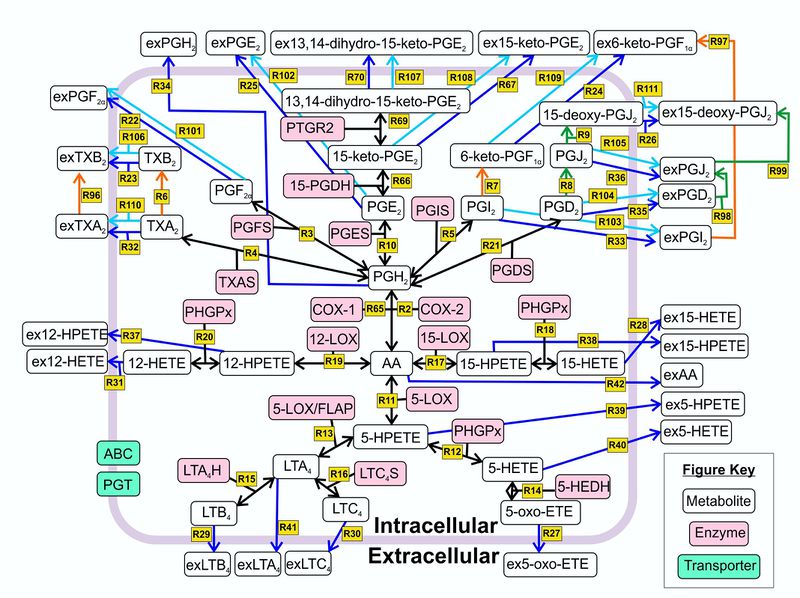
| − | [[ | + | [[Image:Lipidnetwork.jpg|none|thumb|800px|A detailed network of the stimulated arachidonic acid cascade in human skin cells. The metabolites are shown in green rectangles and the enzymes are shown in blue rectangles. The numbered arrows between the metabolites represent the reaction number. The arrows in bold represent the enzyme catalysed reactions, the normal arrows represent non-enzyme catalysed reactions. The degradation and extracellular transport reactions are not shown for simplification purposes. For the full names of the metabolites and enzymes please refere to the abbreviations list.]] |
== Reactions == | == Reactions == | ||
| Line 15: | Line 12: | ||
{|width ="100%" | {|width ="100%" | ||
| | | | ||
| − | |||
| − | |||
| − | |||
'''''Cycloxygenase Reactions''''' | '''''Cycloxygenase Reactions''''' | ||
* [[Transformation of AA to PGH2 |Transformation of AA to PGH<sub>2</sub>, by COX-2 (R2)]] | * [[Transformation of AA to PGH2 |Transformation of AA to PGH<sub>2</sub>, by COX-2 (R2)]] | ||
| Line 24: | Line 18: | ||
* [[Transformation of PGH2 to TXA2 |Transformation of PGH<sub>2</sub> to TXA<sub>2</sub> (R4)]] | * [[Transformation of PGH2 to TXA2 |Transformation of PGH<sub>2</sub> to TXA<sub>2</sub> (R4)]] | ||
* [[Transformation of PGH2 to PGI2 |Transformation of PGH<sub>2</sub> to PGI<sub>2</sub> (R5)]] | * [[Transformation of PGH2 to PGI2 |Transformation of PGH<sub>2</sub> to PGI<sub>2</sub> (R5)]] | ||
| − | |||
* [[Transformation of PGH2 to PGE2 |Transformation of PGH<sub>2</sub> to PGE<sub>2</sub> (R10)]] | * [[Transformation of PGH2 to PGE2 |Transformation of PGH<sub>2</sub> to PGE<sub>2</sub> (R10)]] | ||
| − | * [[Transformation of PGE2 to 15-Keto-PGE2 |Transformation of | + | * [[Transformation of PGH2 to PGD2 |Transformation of PGH<sub>2</sub> to PGD<sub>2</sub> (R21)]] |
| − | * [[Transformation of 15-Keto-PGE2 to 13,14-Dihydro-15-Keto-PGE2|Transformation of 15- | + | * [[Transformation of PGE2 to 15-Keto-PGE2 |Transformation of PGE<sub>2</sub> to 15-Keto-PGE<sub>2</sub> (R66)]] |
| + | * [[Transformation of 15-Keto-PGE2 to 13,14-Dihydro-15-Keto-PGE2|Transformation of 15-keto-PGE<sub>2</sub> to 13,14-dihydro-15-keto-PGE<sub>2</sub> (R69) | ||
]] | ]] | ||
'''''Spontaneous Reactions''''' | '''''Spontaneous Reactions''''' | ||
| − | * [[Transformation of TXA2 to TXB2 |Transformation of TXA<sub>2</sub> to TXB<sub>2</sub> (R6)]] | + | * [[Transformation of TXA2 to TXB2 |Transformation of TXA<sub>2</sub> to TXB<sub>2</sub> (R6/R96)]] |
| − | * [[Transformation of PGI2 to 6-K-PGF1α |Transformation of PGI<sub>2</sub> to 6-keto-PGF<sub>1α</sub> (R7)]] | + | * [[Transformation of PGI2 to 6-K-PGF1α |Transformation of PGI<sub>2</sub> to 6-keto-PGF<sub>1α</sub> (R7/R97)]] |
| − | * [[Transformation of PGD2 to PGJ2 |Transformation of PGD<sub>2</sub> to PGJ<sub>2</sub> (R8)]] | + | * [[Transformation of PGD2 to PGJ2 |Transformation of PGD<sub>2</sub> to PGJ<sub>2</sub> (R8/R98)]] |
| − | * [[Transformation of PGJ2 to 15-D-PGJ2 |Transformation of PGJ<sub>2</sub> to 15-deoxy-PGJ<sub>2</sub> (R9)]] | + | * [[Transformation of PGJ2 to 15-D-PGJ2 |Transformation of PGJ<sub>2</sub> to 15-deoxy-PGJ<sub>2</sub> (R9/R99)]] |
| | | | ||
'''''Lipoxygenase Reactions''''' | '''''Lipoxygenase Reactions''''' | ||
| Line 41: | Line 35: | ||
* [[Transformation of AA to 15-HPETE |Transformation of AA to 15-HPETE (R17)]] | * [[Transformation of AA to 15-HPETE |Transformation of AA to 15-HPETE (R17)]] | ||
* [[Transformation of 5-HPETE to 5-HETE|Transformation of 5-HPETE to 5-HETE (R12)]] | * [[Transformation of 5-HPETE to 5-HETE|Transformation of 5-HPETE to 5-HETE (R12)]] | ||
| − | * [[Transformation of 12-HPETE to 12-HETE|Transformation of 12-HPETE to 12-HETE]] | + | * [[Transformation of 12-HPETE to 12-HETE|Transformation of 12-HPETE to 12-HETE (R20)]] |
* [[Transformation of 15-HPETE to 15-HETE|Transformation of 15-HPETE to 15-HETE (R18)]] | * [[Transformation of 15-HPETE to 15-HETE|Transformation of 15-HPETE to 15-HETE (R18)]] | ||
* [[Transformation of 5-HETE to 5-OXO-ETE|Transformation of 5-HETE to 5-oxo-ETE (R14)]] | * [[Transformation of 5-HETE to 5-OXO-ETE|Transformation of 5-HETE to 5-oxo-ETE (R14)]] | ||
| Line 47: | Line 41: | ||
* [[Transformation of LTA4 to LTB4|Transformation of LTA<sub>4</sub> to LTB<sub>4</sub> (R15)]] | * [[Transformation of LTA4 to LTB4|Transformation of LTA<sub>4</sub> to LTB<sub>4</sub> (R15)]] | ||
* [[Transformation of LTA4 to LTC4|Transformation of LTA<sub>4</sub> to LTC<sub>4</sub> (R16)]] | * [[Transformation of LTA4 to LTC4|Transformation of LTA<sub>4</sub> to LTC<sub>4</sub> (R16)]] | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| | | | ||
'''''Transporter Reactions''''' | '''''Transporter Reactions''''' | ||
| − | * [[ATP-Binding Cassette Transporters|Eicosanoid Export]] | + | * [[ATP-Binding Cassette Transporters|Eicosanoid Export (R22-R42)]] |
| − | * [[Prostaglandin Transporter|Prostaglandin Export]] | + | * [[Prostaglandin Transporter|Prostaglandin Export (R96 -R111)]] |
'''''Degradation Reactions''''' | '''''Degradation Reactions''''' | ||
| − | * [[Degradation Pathways|Degradation Pathways]] | + | * [[Degradation Pathways|Degradation Pathways (R44-64, R68, R71-R94)]] |
| + | '''''Event Reactions''''' | ||
| + | * [[AA Release Event |AA Release Event (R95)]] | ||
| + | * [[COX-2 Induction Event |COX-2 Induction Event (R112)]] | ||
|} | |} | ||
| − | == Model | + | == Model Details == |
| + | * [[Model Constraints|Model Constraints]] | ||
| + | * [[Simulation Details|Simulation Details]] | ||
| + | * [[Model Scripts|Model Scripts]] | ||
| + | * [[Common equations|Equations]] | ||
| + | * [[Abbreviations List|Abbreviations List]] | ||
| − | + | == Application == | |
| − | + | * [[HaCaT keratinocytes|HaCaT keratinocytes]] | |
| − | + | * [[46BR.1N Fibroblasts|46BR.1N Fibroblasts]] | |
| − | |||
| − | * [[ | ||
| − | * [[ | ||
| − | == | + | == Future Areas of Research == |
| + | * [[Network Expansion|Network Expansion]] | ||
== References == | == References == | ||
<references/> | <references/> | ||
Latest revision as of 08:33, 21 August 2019
Contents
Description of the Model
Computational models have been demonstrated as useful tools to gain a better understanding of networks, aid rational decision-making and support research across the biosciences[1][2][3]. Due to the complexity of the AA cascade, predicting the outcome of the network is difficult due to the collaborative action of numerous species (i.e. metabolites, proteins, signalling cascades). In addition, the AA cascade is ubiquitous and is implicated in numerous inflammatory diseases such as cancer, atherosclerosis and asthma [4][5] [6][7]; as a result, the production of eicosanoids is the targets of several pharmacological interventions.Therefore, there is a need to utilise a systems-level approach to further investigate the bioactive AA cascade. This can be achieved by building upon previous models of the AA cascade [8][9][10] and creating a new model which addresses the gaps in the literature such as an extensive network topology, the use of experimental parameters, documentation of parameter sources, integration of data from multiple platforms and application beyond a single cell type.
Furthermore, there is a need to incorporate novel approaches to the parameterisation and simulation of metabolic models, in order to quantify the uncertainty associated with literature data and indicate the robustness of each prediction [11]. The main aim of this project is to construct, analyse and validate a predictive and adaptable metabolic model of the AA cascade. To use this model to investigate how the concentration of metabolites change over time following internal/external perturbations, we need to utilise a kinetic deterministic modelling approach. This work will build upon the previous models of the AA cascade and support researchers across the biosciences to gain a better understanding of pathways by incorporating means of adapting the model to represent different cell types. We will demonstrate the application of this work in immortalised human cell lines, HaCaT keratinocytes and 46BR.1N fibroblasts as they are shown to produce different eicosanoids, availabilities of AA, protein profiles and response to inflammatory stimuli.
Reactions
Literature information surrounding every reaction within the model are described on the following pages:
Model Details
Application
Future Areas of Research
References
- ↑ Yang, K. Ma, W. Liang, H. Ouyang, Q. Tang, C. Lai, L., Dynamic simulations on the arachidonic acid metabolic network, PLoS Comput Biol (2007), 3, e55
- ↑ Gupta, S. Maurya, M. R. Stephens, D. L. Dennis, E. A. Subramaniam, S., An integrated model of eicosanoid metabolism and signalling based on lipidomics flux analysis, Biophys J (2009), 96, 4542-51.
- ↑ Dobovisek, A. Fajmut, A. Brumen, M., Role of expression of prostaglandin synthases 1 and 2 and leukotriene C4 synthase in aspirin-intolerant asthma: a theoretical study, J Pharmacokinet Pharmacodyn (2011), 38, 261-78.
- ↑ Bishop-Bailey, D. Calatayud, S. Warner, T. D. Hla, T. Mitchell, J. A., Prostaglandins and the regulation of tumor growth, J Environ Pathol Toxicol Oncol (2002), 21, 93-101
- ↑ von Schacky, C., Omega-3 fatty acids in cardiovascular disease - an uphill battle., Prostaglandins Leukot Essent Fatty Acids (2015), 92, 41-7.
- ↑ Claar, D. Hartert, T. V. Peebles, R. S., Jr., The role of prostaglandins in allergic lung inflammation and asthma, Expert Rev Respir Med (2015), 9, 55-72.
- ↑ Albanesi, C. Pastore, S., Pathobiology of chronic inflammatory skin diseases: interplay between keratinocytes and immune cells as a target for anti-inflammatory drugs, Curr Drug Metab (2010), 11, 210-27.
- ↑ Yang, K. Ma, W. Liang, H. Ouyang, Q. Tang, C. Lai, L., Dynamic simulations on the arachidonic acid metabolic network, PLoS Comput Biol (2007), 3, e55
- ↑ Gupta, S. Maurya, M. R. Stephens, D. L. Dennis, E. A. Subramaniam, S., An integrated model of eicosanoid metabolism and signalling based on lipidomics flux analysis, Biophys J (2009), 96, 4542-51.
- ↑ Dobovisek, A. Fajmut, A. Brumen, M., Role of expression of prostaglandin synthases 1 and 2 and leukotriene C4 synthase in aspirin-intolerant asthma: a theoretical study, J Pharmacokinet Pharmacodyn (2011), 38, 261-78.
- ↑ Tsigkinopoulou, A. Hawari, A. Uttley, M. Breitling, R., Defining informative priors for ensemble modeling in systems biology, Nat Protoc (2018), 13, 2643-2663.