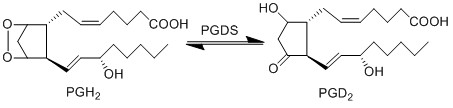
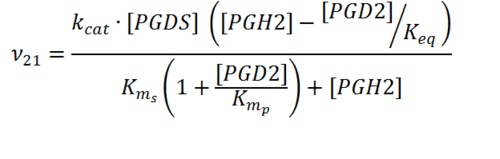Transformation of PGH2 to PGD2
The isomerisation of PGH2 to PGD2 can be performed by prostaglandin D synthase (PGDS) and prostaglandin F synthase (PGFS). PGDS yields a hydroxyl group at C9 and a ketone group at C11. Two isoforms of PGDS have been found in humans, the lipocalin isoform, L-PGDS, does not require glutathione for catalysis whereas the hemopoietic type, H-PGDS, does require glutathione.
Contents
Reaction
Chemical equation
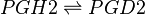
Rate equation
Enzyme Parameters
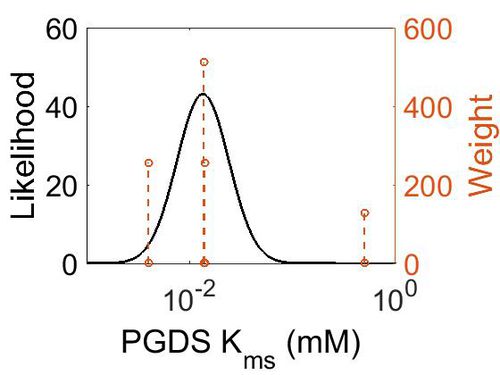
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 0.0138 | 
|
Human | Expression Vector: Human Cell
Enzyme: PGDS pH: 8 Temperature: 25 |
512 | [1] |
| 4.00E-03 | 
|
Human | Expression Vector: Cerebrospinal Fluid
Enzyme: PGDS pH:10 Temperature: Unspecified |
256 | [2] |
| 0.50 | 
|
Human | Expression Vector: E. Coli.
Enzyme: PGDS pH:6.5 Temperature: Unspecified |
128 | [3] |
| 1.40E-02 | 
|
Rat | Expression Vector: Cerebrospinal Fluid
Enzyme: PGDS pH:7 Temperature: 25 |
256 | [4] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.35E-02 | 3.78E+00 | -3.44E+00 | 9.29E-01 |
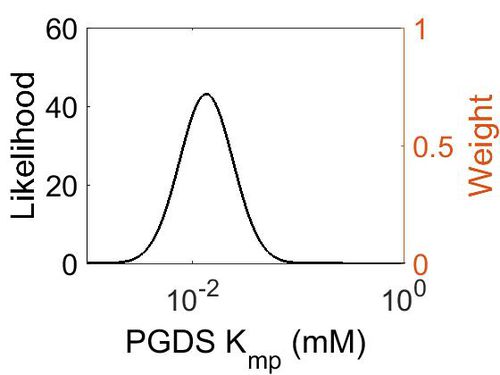
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.30E-02 | -4.02E+00 | 5.69E-01 |
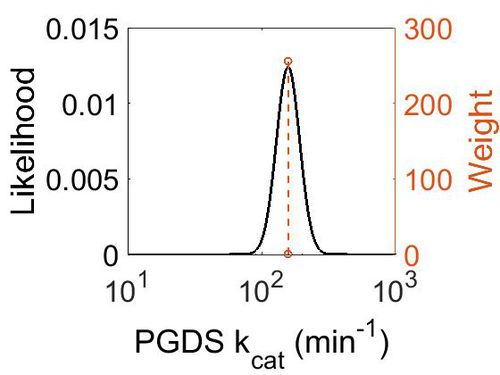
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 158.4 | per minute | Human | Expression Vector: Human Cell
Enzyme: PGDS pH: 8 Temperature: 25 |
256 | [5] |
| 1302 | per minute | Human | Expression Vector: E. Coli.
Enzyme: PGDS pH:6.5 Temperature: Unspecified |
128 | [6] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.58E+02 | 1.50E+00 | 5.10E+00 | 2.00E-01 |
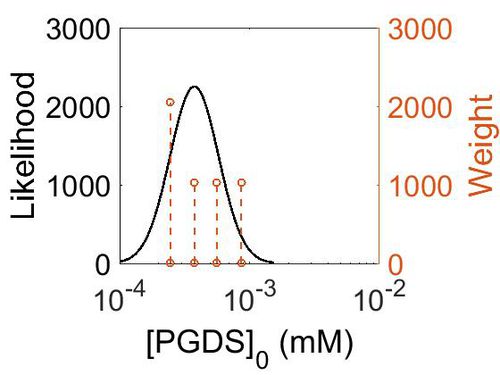
Enzyme concentrations
To convert the enzyme concentration from ppm to mM, the following equation was used.
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 156 | 
|
Human | Expression Vector: Pancreas
Enzyme: PGDS pH: 7.5 Temperature: 37 °C |
1024 | [7] |
| 101 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: PGDS pH: 7.5 Temperature: 37 °C |
1024 | [8] |
| 67.9 | 
|
Human | Expression Vector: Esophagus
Enzyme: PGDS pH: 7.5 Temperature: 37 °C |
1024 | [7] |
| 44.5 | 
|
Human | Expression Vector:Skin
Enzyme: PGDS pH: 7.5 Temperature: 37 °C |
2048 | [8] |
| Mode (ppm) | Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|---|
| 6.76E+01 | 3.74E-04 | 1.64E+00 | 4.41E+00 | 4.49E-01 |
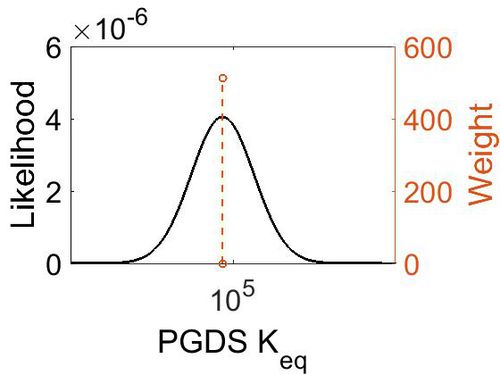
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
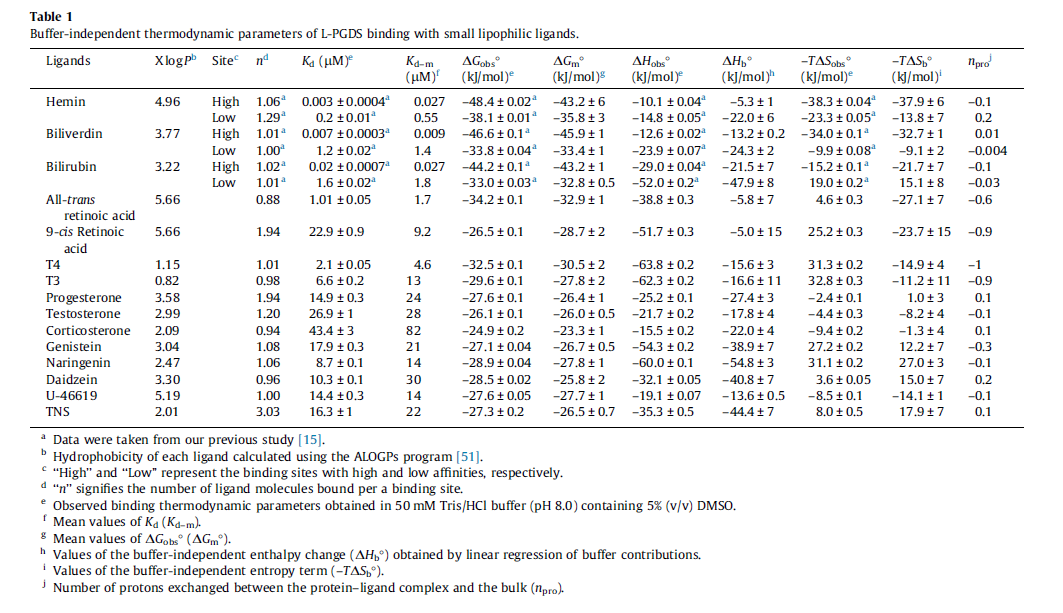
|
|
Human | Expression Vector: E. Coli
Enzyme: L-PGDS pH: 8 Temperature: 25 |
64 | [9] |
| 5.72 | kcal/mol | Not stated | Estimated
Enzyme: PGDS Substrate: Arachidonate Product: PGD2 pH: 7.3 ionic strength: 0.25 |
64 | [10] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 7.46E+04 | 1.00E+01 | 1.20E+01 | 8.90E-01 |
References
- ↑ Zhou Y. , "Structure-function analysis of human l-prostaglandin D synthase bound with fatty acid molecules." FASEB J. 2010 Dec;24(12):4668-77. doi: 10.1096/fj.10-164863. Epub 2010 Jul 28.
- ↑ Watanabe K. , "Identification of beta-trace as prostaglandin D synthase."Biochem Biophys Res Commun. 1994 Sep 15;203(2):1110-6.
- ↑ Pinzar E , "Structural basis of hematopoietic prostaglandin D synthase activity elucidated by site-directed mutagenesis." J Biol Chem. 2000 Oct 6;275(40):31239-44.
- ↑ Urade Y. , "Purification and characterization of rat brain prostaglandin D synthetase." J Biol Chem. 1985 Oct 15;260(23):12410-5.
- ↑ Zhou Y. , "Structure-function analysis of human l-prostaglandin D synthase bound with fatty acid molecules. FASEB J. 2010 Dec;24(12):4668-77.
- ↑ [https://www.ncbi.nlm.nih.gov/pubmed/10871602 Pinzar E , "Structural basis of hematopoietic prostaglandin D synthase activity elucidated by site-directed mutagenesis. J Biol Chem. 2000 Oct 6;275(40):31239-44. ]
- ↑ 7.0 7.1 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ 8.0 8.1 M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ [http://ac.els-cdn.com/S0014579314000982/1-s2.0-S0014579314000982-main.pdf?_tid=7f323a24-6543-11e6-82ac-00000aab0f01&acdnat=1471525305_289e200c1e492c5fa693179a5396fd82 Kume S., "Fine-tuned broad binding capability of human lipocalin-type prostaglandin D synthase for various small lipophilic ligands FEBS Letters Volume 588, Issue 6, 18 March 2014, Pages 962–969]
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471