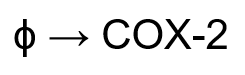COX-2 Induction Event
The induced expression of COX-2. As this process is as a result of complex transcription and translation processes, this reaction was simplified.
In this reaction there are parameters which can be manually adapted depending on the in silico experiment. This means that the value of these parameters can change throughout the simulation unlike other parameters which, once assigned, remain fixed for the entire model variant. These parameters have no molecular meaning but were included in order to allow the user to manually adapt the model to represent different scenarios, such as different treatments.
Reaction
Rate equation
Parameters
| Parameter Name | Parameter Shorthand | Initial Value | Justification |
|---|---|---|---|
| Doubling time of the COX-2 event | COX2DT | 180 min | This value was upon the unpublished western blot data in (Kiezel-Tsugunova, 2017). |
| Maximum concentration of COX-2 | C3 | 0 mM | The adapted value of this parameter depends on the scenario (Table *). This value changes 240 min into the simulation for the UVR scenarios. |
| Decay switch | C4 | 0 | If required the induced expression of COX-2 can decrease over time by amending this value from 0 to 1. This was set to 0 in all scenarios. |
| Half-life of the COX-2 event | COX2HL | 0 min | This value was arbitrarily set at 0 min. However, no simulations required COX-2 release to decrease over time. |
| In silico experiment | HaCaT keratinocyte + A23187 | 46BR.1N fibroblast +A23187 | HaCaT keratinocyte + Indomethacin and A23187 | 46BR.1N fibroblast + Indomethacin and A23187 | HaCaT keratinocyte + ATP | 46BR.1N fibroblast + ATP | HaCaT keratinocyte + UVR | 46BR.1N fibroblast + UVR |
|---|---|---|---|---|---|---|---|---|
| COX-1 (mM) at 0h | x | x | 1.00 x10-28 | 1.00 x10-28 | x | x | x | x |
| COX-2 (mM) at 0h | x | x | 1.00 x10-28 | 1.00 x10-28 | x | x | 1.00 x10-28 | 1.00 x10-28 |
| COX-2 (mM) at 6h | x | x | x | x | x | x | 2.27 x10-2 | 2.27 x10-2 |