Difference between revisions of "Welcome to the In-Silico Model of butyrolactone regulation in Streptomyces coelicolor"
Marcbiarnesc (talk | contribs) (→About the Project) |
Marcbiarnesc (talk | contribs) (→About the Project) |
||
| Line 4: | Line 4: | ||
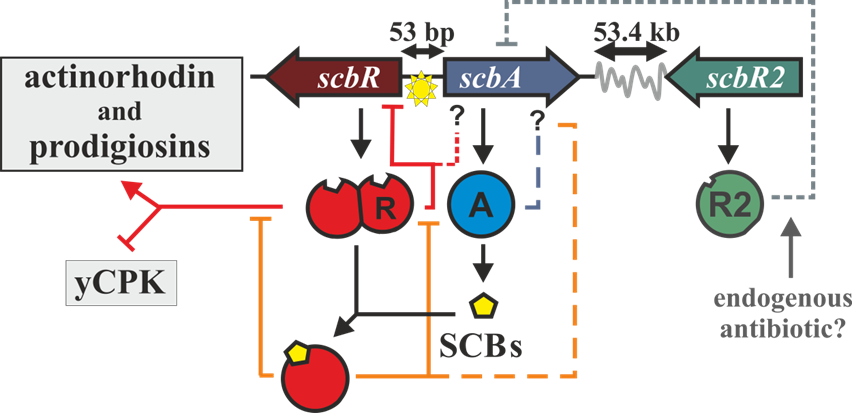
The ''scbA'' gene is presumed to catalyse the condensation of dihydroxyacetone phosphate with a beta-ketoacid to produce three different butyrolactones (SCB1, SCB2 and SCB3. Hereafter named as SCBs). On the other hand, ''scbR'' is a TetR-like DNA-binding protein known to regulate its own transcription and that of ''scbA'' and to directly regulate production of a cryptic metabolite and indirectly regulate production of blue pigmented actinorhodin (Act) and prodigiosins (e.g. undecylprodigiosin, Red). These genes are divergently encoded and their promoter regions overlap 53bp. | The ''scbA'' gene is presumed to catalyse the condensation of dihydroxyacetone phosphate with a beta-ketoacid to produce three different butyrolactones (SCB1, SCB2 and SCB3. Hereafter named as SCBs). On the other hand, ''scbR'' is a TetR-like DNA-binding protein known to regulate its own transcription and that of ''scbA'' and to directly regulate production of a cryptic metabolite and indirectly regulate production of blue pigmented actinorhodin (Act) and prodigiosins (e.g. undecylprodigiosin, Red). These genes are divergently encoded and their promoter regions overlap 53bp. | ||
| − | [[File:Picture1.png]] | + | [[File:Picture1.png]] |
Transcription analysis have shown that both genes are mainly active during transition from logaritmic growth to stationary phase. It is presumed that SCBs slowly accumulate into the media and upon reaching a concentration threshold promote a coordinated switch-like transition to antibiotic production by binding to ScbR. However, the mechanism of this network is not fully defined, although several alternative scenarios have been proposed. In 2008, Mehra et al ('''''ref''''') proposed a deterministic model involving a putative ScbR-ScbA complex. More recently, Chatterjee et al ('''''ref''''') proposed that the promoter overlap between ''scbA'' and ''scbR'' is the sole driving force of the precise switch-like transition. | Transcription analysis have shown that both genes are mainly active during transition from logaritmic growth to stationary phase. It is presumed that SCBs slowly accumulate into the media and upon reaching a concentration threshold promote a coordinated switch-like transition to antibiotic production by binding to ScbR. However, the mechanism of this network is not fully defined, although several alternative scenarios have been proposed. In 2008, Mehra et al ('''''ref''''') proposed a deterministic model involving a putative ScbR-ScbA complex. More recently, Chatterjee et al ('''''ref''''') proposed that the promoter overlap between ''scbA'' and ''scbR'' is the sole driving force of the precise switch-like transition. | ||
Revision as of 16:35, 25 September 2015
Contents
About the Project
Streptomyces are Gram-positive soil-dwelling bacteria, which are known for being prolific sources of secondary metabolites, many of which have medical interest (e.g.: streptomycin, cloramphenicol or kanamycin). Recent genome sequencing, and posterior bioinformatic analysis, using tools such as antiSMASH [[1]], have shown the presence of putative cryptic secondary metabolite-producing clusters. One way Streptomyces can coordinate secondary metabolite production is through the use of small diffusible molecules, known as γ-butyrolactones. In the model organism Streptomyces coelicolor the butyrolactone regulatory system involves a synthase (scbA) and a butyrolactone receptor scbR.
The scbA gene is presumed to catalyse the condensation of dihydroxyacetone phosphate with a beta-ketoacid to produce three different butyrolactones (SCB1, SCB2 and SCB3. Hereafter named as SCBs). On the other hand, scbR is a TetR-like DNA-binding protein known to regulate its own transcription and that of scbA and to directly regulate production of a cryptic metabolite and indirectly regulate production of blue pigmented actinorhodin (Act) and prodigiosins (e.g. undecylprodigiosin, Red). These genes are divergently encoded and their promoter regions overlap 53bp.
Transcription analysis have shown that both genes are mainly active during transition from logaritmic growth to stationary phase. It is presumed that SCBs slowly accumulate into the media and upon reaching a concentration threshold promote a coordinated switch-like transition to antibiotic production by binding to ScbR. However, the mechanism of this network is not fully defined, although several alternative scenarios have been proposed. In 2008, Mehra et al (ref) proposed a deterministic model involving a putative ScbR-ScbA complex. More recently, Chatterjee et al (ref) proposed that the promoter overlap between scbA and scbR is the sole driving force of the precise switch-like transition.
The aim of this project is the design and analysis of a stochastic and parameter uncertainty-aware model which describes the butyrolactone regulatory system and allows reliable predictions of its behaviour. The complete elucidation of this system could potentially lead to the design of robust and sensitive systems as orthologous regulatory circuits in synthetic biology and biotechnology.
Description of the Model
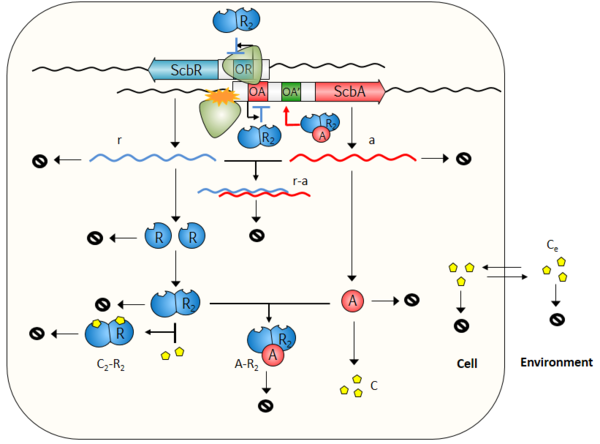
Several alternative scenarios for the mechanism of action of the GBL system have been previously proposed. Our aim is to create a unified model which will include them all and enable their parallel or combined analysis. The scenarios investigated are the following:
- The formation of a complex between proteins ScbA and ScbR, which relieves the self-repression of ScbR and the repression of ScbA by ScbR, while at the same time activates the production of ScbA and in turn of SCB1. [1]
- The effect of transcriptional interference (collisions between the elongating RNAPs which leads to transcriptional termination) due to the overlap of the two genes' promoter regions by 53 bp and by the convergent transcription of the two genes. This results in a decrease in expression of full-length RNAs from both promoters and production of truncated RNAs. [2] [3]
- The antisense effect conferred by convergent transcription of the ScbR and ScbA genes. In this case, transcripts with a segment of complementary sequence may lead to interactions between sense-antisense full length transcripts of the two genes, thus leading to the formation of a fast degrading complex of the two mRNAs and inhibition of translation. [2]
The model consists of two compartments, the Cell and the Environment. A schematic description of the model is provided below. More information can be obtained by clicking on each reaction arrow in the figure.
Species
| Name | Description | Compartment |
|---|---|---|
| OR | Promoter of ScbR gene | Cell |
| OA | Promoter of ScbA gene | Cell |
| OA' | Alternative promoter for activation of ScbA gene by the AR2 complex | Cell |
| OR-R2 | Complex of R protein and promoter of ScbR gene | Cell |
| OA-R2 | Complex of R protein and promoter of ScbA gene | Cell |
| OA'-AR2 | Alternative promoter for activation of ScbA gene by the AR2 complex | Cell |
| r | mRNA transcript of ScbR gene | Cell |
| a | mRNA transcript of ScbA gene | Cell |
| r-a | complex of full length ScbR and ScbA mRNAs due to antisense effect | Cell |
| R | ScbR protein | Cell |
| R2 | ScbR homo-dimer | Cell |
| A | ScbA protein | Cell |
| C | SCB1 (γ-butyrolactone) | Cell |
| AR2 | ScbA-ScbR complex | Cell |
| S | Glycerol derivative and b-keto acid derivative precursors | Cell |
| C2R2 | SCB1-ScbR complex | Cell |
| Ce | Extracellular SCB1 (γ-butyrolactone) | Environment |
Reactions
The reactions per compartment are the following:
|
Cell |
Cell-Environment
Environment |
Parameter Overview
References
- ↑ S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 2.0 2.1 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ E. Takano. γ-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Current Opinion in Microbiology, 9(3):287–294, 2006.