Difference between revisions of "Welcome to the In-Silico Model of Cutaneous Lipids Wiki"
(→Reactions) |
(→Reactions) |
||
| Line 43: | Line 43: | ||
* [[Transformation of 12-HPETE to 12-HETE|Transformation of 12-HPETE to 12-HETE]] | * [[Transformation of 12-HPETE to 12-HETE|Transformation of 12-HPETE to 12-HETE]] | ||
* [[Transformation of 15-HPETE to 15-HETE|Transformation of 15-HPETE to 15-HETE]] | * [[Transformation of 15-HPETE to 15-HETE|Transformation of 15-HPETE to 15-HETE]] | ||
| − | * [[Transformation of 5-HETE to 5-OXO-ETE|Transformation of 5-HETE to 5- | + | * [[Transformation of 5-HETE to 5-OXO-ETE|Transformation of 5-HETE to 5-oxo-ETE (R14)]] |
* [[Transformation of 5-HPETE to LTA4|Transformation of 5-HPETE to LTA<sub>4</sub> (R13)]] | * [[Transformation of 5-HPETE to LTA4|Transformation of 5-HPETE to LTA<sub>4</sub> (R13)]] | ||
* [[Transformation of LTA4 to LTB4|Transformation of LTA4 to LTB4]] | * [[Transformation of LTA4 to LTB4|Transformation of LTA4 to LTB4]] | ||
Revision as of 12:00, 15 May 2019
The skin is the largest structure in the body and acts as the ultimate barrier to our environment. The aim of this project is to comprehend how the lipid network within the skin responds in physiopathological states such as inflammation (sunburn, atopic dermatitis, psoriasis). Little is known about the regulatory relationships within the cutaneous lipid signalling networks, and there is a distinct lack of information about how these networks interact between compartments (the different layers of the skin).
Contents
Description of the Model
We aim to create a quantitative computational model of the cutaneous lipid signalling networks, including variables such as multiple lipid substrates and enzyme isoforms. Model construction will be guided by literature studies, combined with new experimental lipidomics data collected in this project. In order to analyse model properties, experimental studies such as altering the systemic availability of fatty acids and the effect upon the anti-inflammatory milieu will be measured. This information will be integrated into the model to obtain estimated kinetic parameter values and also to validate and refine the overall model topology. The resulting mathematical model will be testable and predictive of the lipid responses during inflammation. The insights obtained will allow for more targeted experiments to be designed in order to better understand skin lipid biology and to aid in the mapping of lipid networks contributing to inflammation. This will ultimately support the design of interventional studies to combat skin disease.
The full list of common acronyms used are listed here.
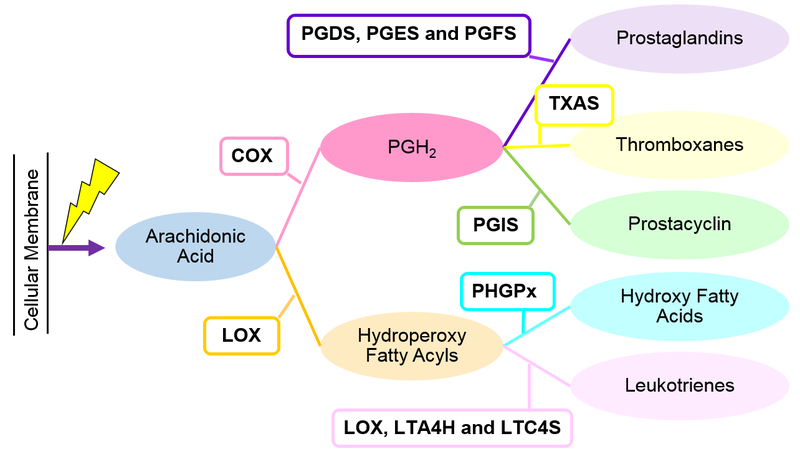
Reactions
Literature information surrounding every reaction within the model are described on the following pages:
Model Parameterisation
- Enzyme Parameters
- Initial Concentrations
- Transport Parameters
- Non-Enzyme Catalysed Reaction Parameters
- Degradation Parameters
- Model Constraints