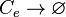Difference between revisions of "Model Overview"
(→List of Model Reactions) |
(→Parameters for model reactions) |
||
| Line 86: | Line 86: | ||
! Distribution | ! Distribution | ||
Mode | Mode | ||
| − | ! | + | ! Spread |
| − | |||
! Location | ! Location | ||
parameter (μ) | parameter (μ) | ||
| Line 98: | Line 97: | ||
|<math> nM </math> | |<math> nM </math> | ||
|0.299 | |0.299 | ||
| − | | | + | |6.8 |
| − | |0. | + | |0.15471 |
| − | |1. | + | |1.1661 |
|- | |- | ||
|<math>k^{-}_{1}, k^{-}_{7}</math> | |<math>k^{-}_{1}, k^{-}_{7}</math> | ||
| Line 106: | Line 105: | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
|0.489 | |0.489 | ||
| − | | | + | |1.98 |
| − | |−0. | + | |−0.37642 |
| − | |0. | + | |0.58225 |
|- | |- | ||
|rowspan="2" |[[Binding of R2 to OA operator |Binding of R<sub>2</sub> to OA operator]] | |rowspan="2" |[[Binding of R2 to OA operator |Binding of R<sub>2</sub> to OA operator]] | ||
| Line 115: | Line 114: | ||
|<math> nM </math> | |<math> nM </math> | ||
|0.299 | |0.299 | ||
| − | | | + | |6.8 |
| − | |0. | + | |0.15471 |
| − | |1. | + | |1.1661 |
|- | |- | ||
|<math>k^{-}_{2},k^{-}_{8}</math> | |<math>k^{-}_{2},k^{-}_{8}</math> | ||
| Line 123: | Line 122: | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
|0.489 | |0.489 | ||
| − | | | + | |1.98 |
| − | |−0. | + | |−0.37642 |
| − | |0. | + | |0.58225 |
|- | |- | ||
|rowspan="2" |[[Binding of R2 to A |Binding of R<sub>2</sub> to A]] | |rowspan="2" |[[Binding of R2 to A |Binding of R<sub>2</sub> to A]] | ||
| Line 131: | Line 130: | ||
|<math>10^{4}−10^{6}</math> | |<math>10^{4}−10^{6}</math> | ||
|<math> nM </math> | |<math> nM </math> | ||
| − | |<math> | + | |<math>9.9 \cdot 10^{4}</math> |
| − | | | + | |10.02 |
| − | | | + | |13.192 |
| − | |1. | + | |1.2977 |
|- | |- | ||
|<math>k^{-}_{3}</math> | |<math>k^{-}_{3}</math> | ||
|<math>38.3-4.46 \cdot 10^{6}</math> | |<math>38.3-4.46 \cdot 10^{6}</math> | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | | | + | |N/A |
| − | | | + | |N/A |
| − | | | + | |9.9423 |
| − | |1. | + | |1.5503 |
|- | |- | ||
|rowspan="2" |[[Binding of R2 to C |Binding of R<sub>2</sub> to C]] | |rowspan="2" |[[Binding of R2 to C |Binding of R<sub>2</sub> to C]] | ||
| Line 148: | Line 147: | ||
|<math>0.0011-10400</math> | |<math>0.0011-10400</math> | ||
|<math> nM </math> | |<math> nM </math> | ||
| − | | | + | |N/A |
| − | | | + | |N/A |
| − | | | + | |5.9389 |
| − | |2. | + | |2.3412 |
|- | |- | ||
|<math>k^{-}_{4}</math> | |<math>k^{-}_{4}</math> | ||
|<math>3 \cdot 10^{-4}-126</math> | |<math>3 \cdot 10^{-4}-126</math> | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.816 |
| − | | | + | |244 |
| − | | | + | |4.2872 |
| − | |2. | + | |2.119 |
|- | |- | ||
|rowspan="2" |[[Binding of A-R2 to OA' operator |Binding of A-R<sub>2</sub> to OA' operator]] | |rowspan="2" |[[Binding of A-R2 to OA' operator |Binding of A-R<sub>2</sub> to OA' operator]] | ||
| Line 166: | Line 165: | ||
|<math> nM </math> | |<math> nM </math> | ||
|0.299 | |0.299 | ||
| − | | | + | |6.8 |
| − | |0. | + | |0.15471 |
| − | |1. | + | |1.1661 |
|- | |- | ||
|<math>k^{-}_{5},k^{-}_{9},k^{-}_{10},k^{-}_{11},k^{-}_{12}</math> | |<math>k^{-}_{5},k^{-}_{9},k^{-}_{10},k^{-}_{11},k^{-}_{12}</math> | ||
| Line 174: | Line 173: | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
|0.489 | |0.489 | ||
| − | | | + | |1.98 |
| − | |−0. | + | |−0.37642 |
| − | |0. | + | |0.58225 |
|- | |- | ||
|[[Synthesis of C]] | |[[Synthesis of C]] | ||
|<math>K_{C}</math> | |<math>K_{C}</math> | ||
| − | |0. | + | |0.00315−85.5 |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.094 |
| − | | | + | |60.49 |
| − | | | + | |0.87914 |
| − | | | + | |1.8017 |
|- | |- | ||
|rowspan="3" |[[Transcription of r]] | |rowspan="3" |[[Transcription of r]] | ||
| Line 192: | Line 191: | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
|0.8346 | |0.8346 | ||
| − | | | + | |3.55 |
| − | |0. | + | |0.63056 |
| − | |0. | + | |0.90076 |
|- | |- | ||
|<math>k_{F}</math> | |<math>k_{F}</math> | ||
|18.2−33 | |18.2−33 | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |20. | + | |20.7 |
| − | |1. | + | |1.34 |
| − | |3. | + | |3.1107 |
| − | |0. | + | |0.28276 |
|- | |- | ||
|<math>k_{onR}</math> | |<math>k_{onR}</math> | ||
| Line 208: | Line 207: | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
|0.867 | |0.867 | ||
| − | | | + | |4.09 |
| − | |0. | + | |0.78694 |
| − | |1. | + | |0.96428 |
| + | |- | ||
| + | |rowspan="3" |[[Transcription of a]] | ||
| + | |<math>\Omega_{A_{basal}}</math> | ||
| + | |0.001−0.2 | ||
| + | |<math> min^{-1} </math> | ||
| + | |0.0037 | ||
| + | |10.93 | ||
| + | |−3.8324 | ||
| + | |1.3259 | ||
|- | |- | ||
| − | |||
|<math>\Omega_{A}</math> | |<math>\Omega_{A}</math> | ||
|0.11−5.65 | |0.11−5.65 | ||
Revision as of 17:24, 23 November 2017
Contents
List of Model Reactions
*1 Reactions taking place only in the scenario which includes the activating complex 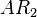
*2 Reactions taking place only in the scenario which includes the antisense RNA interactions
| Reactions |
|---|
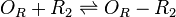
|
Parameters for model reactions
| Reaction | Parameter | Value | Units | Distribution
Mode |
Spread | Location
parameter (μ) |
Scale
parameter (σ) |
|---|---|---|---|---|---|---|---|
| Binding of R2 to OR operator | 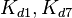
|
0.005−5.8 | 
|
0.299 | 6.8 | 0.15471 | 1.1661 |
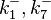
|
0.09−0.846 | 
|
0.489 | 1.98 | −0.37642 | 0.58225 | |
| Binding of R2 to OA operator | 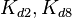
|
0.005−5.8 | 
|
0.299 | 6.8 | 0.15471 | 1.1661 |
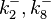
|
0.09−0.846 | 
|
0.489 | 1.98 | −0.37642 | 0.58225 | |
| Binding of R2 to A | 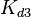
|
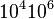
|

|
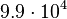
|
10.02 | 13.192 | 1.2977 |

|
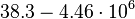
|

|
N/A | N/A | 9.9423 | 1.5503 | |
| Binding of R2 to C | 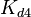
|
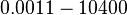
|

|
N/A | N/A | 5.9389 | 2.3412 |
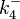
|
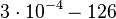
|

|
0.816 | 244 | 4.2872 | 2.119 | |
| Binding of A-R2 to OA' operator | 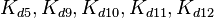
|
0.005−5.8 | 
|
0.299 | 6.8 | 0.15471 | 1.1661 |
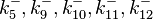
|
0.09−0.846 | 
|
0.489 | 1.98 | −0.37642 | 0.58225 | |
| Synthesis of C | 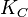
|
0.00315−85.5 | 
|
0.094 | 60.49 | 0.87914 | 1.8017 |
| Transcription of r | 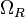
|
0.16−8.24 | 
|
0.8346 | 3.55 | 0.63056 | 0.90076 |
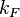
|
18.2−33 | 
|
20.7 | 1.34 | 3.1107 | 0.28276 | |
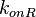
|
0.0518-14.5 | 
|
0.867 | 4.09 | 0.78694 | 0.96428 | |
| Transcription of a | 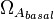
|
0.001−0.2 | 
|
0.0037 | 10.93 | −3.8324 | 1.3259 |
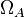
|
0.11−5.65 | 
|
0.57 | 12.5 | 0.354505 | 0.95491 | |
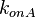
|
0.039-8.82 | 
|
0.587 | 15 | 0.47762 | 1.0048 | |
| Antisense interaction between r and a | 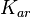
|
0.4−89 | 
|
7.8 | 52.8 | 3.7933 | 1.3186 |
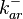
|
0.006−6 | 
|
0.22 | 23.2 | −0.24716 | 1.1192 | |
| Translation of R | 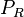
|
0.165−5.86 | 
|
0.74 | 10.3 | 0.50964 | 0.89756 |
| Translation of A | 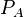
|
0.11−4 | 
|
0.51 | 10.3 | 0.13413 | 0.89936 |
| Formation of homo-dimer R2 | 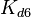
|
2.58-42.9 | 
|
30 | 30 | 4.8018 | 1.1835 |
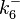
|
0.144-51.2 | 
|
2 | 30 | 2.094 | 1.1835 | |
| Degradation of r | 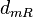
|
0.03−0.365 | 
|
0.14 | 4.24 | −1.5762 | 0.62077 |
| Degradation of R | 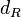
|
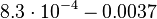
|

|
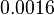
|
2.5 | −6.2752 | 0.4285 |
| Degradation of R2 | 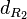
|
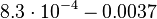
|

|
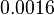
|
2.5 | −6.2752 | 0.4285 |
| Degradation of a | 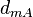
|
0.03−0.365 | 
|
0.14 | 4.24 | −1.5762 | 0.62077 |
| Degradation of A | 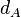
|
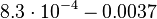
|

|
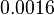
|
2.5 | −6.2752 | 0.4285 |
| Degradation of C | 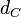
|
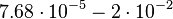
|

|
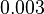
|
25.8 | −4.4205 | 1.1461 |
| Degradation of C2-R2 | 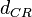
|
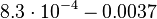
|

|
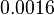
|
2.5 | −6.2752 | 0.4285 |
| Degradation of A-R2 | 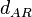
|
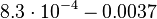
|

|
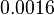
|
2.5 | −6.2752 | 0.4285 |
| Degradation of r-a | 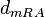
|
0.35−69.3 | 
|
0.739 | 200.4 | 2.2813 | 1.6074 |
| Diffusion of C and Ce | 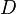
|
0.1−24 | 
|
2.8 | 18.2 | 2.1457 | 1.0564 |
| Degradation of Ce | 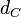
|
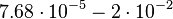
|

|
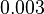
|
25.8 | −4.4205 | 1.1461 |
| Cellular growth | Failed to parse (PNG conversion failed; check for correct installation of latex and dvipng (or dvips + gs + convert)): μ | 0.0004−0.0107 | 
|
0.0022 | 7.5 | −5.4517 | 0.80808 |
Parameters for cell colony growth
| Parameter | Value | Units |
|---|---|---|
| Failed to parse (PNG conversion failed; check for correct installation of latex and dvipng (or dvips + gs + convert)): τ | 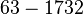
|
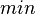
|
| Failed to parse (PNG conversion failed; check for correct installation of latex and dvipng (or dvips + gs + convert)): λ | 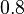
|

|
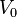
|
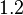
|
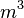
|
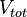
|
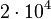
|
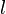
|
Initial concentrations of species
| Species | Initial concentration | Units | Compartment |
|---|---|---|---|
| OR | 0.9592 | 
|
Cell |
| OA | 0.9592 | 
|
Cell |
| OR-R2 | 0 | 
|
Cell |
| OA-R2 | 0 | 
|
Cell |
| OA'-AR2 | 0 | 
|
Cell |
| r | 0 | 
|
Cell |
| a | 0 | 
|
Cell |
| r-a | 0 | 
|
Cell |
| R | 0 | 
|
Cell |
| R2 | 0 | 
|
Cell |
| A | 0 | 
|
Cell |
| C | 0 | 
|
Cell |
| AR2 | 0 | 
|
Cell |
| C2R2 | 0 | 
|
Cell |
| Ce | 0 | 
|
Environment |


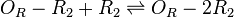
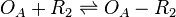
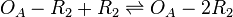
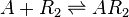 *1
*1
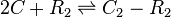
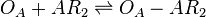 *1
*1
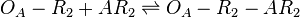 *1
*1
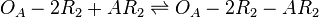 *1
*1
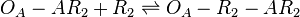 *1
*1
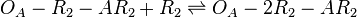 *1
*1
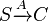
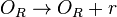
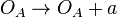
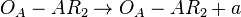 *1
*1
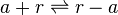 *2
*2
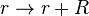
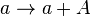
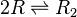
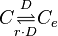
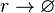
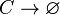
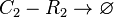
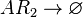 *1
*1
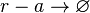 *2
*2