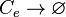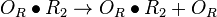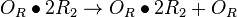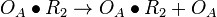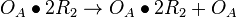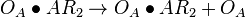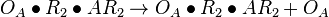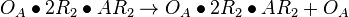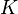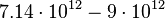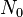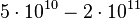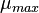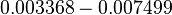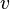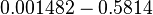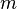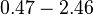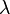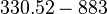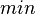Difference between revisions of "Model Overview"
(→Parameters for model reactions) |
|||
| (86 intermediate revisions by the same user not shown) | |||
| Line 3: | Line 3: | ||
default [[Welcome to the In-Silico Model of γ-butyrolactone regulation in Streptomyces coelicolor|Go back to overview]] | default [[Welcome to the In-Silico Model of γ-butyrolactone regulation in Streptomyces coelicolor|Go back to overview]] | ||
</imagemap> | </imagemap> | ||
| + | |||
| + | == Model scripts == | ||
| + | |||
| + | The model scripts for the different scenarios can be found in pdf form here: [[File:GBL_Models.pdf]] | ||
| + | |||
| + | == List of Model Reactions == | ||
| + | |||
| + | <sup>*1</sup> Reactions taking place only in the scenarios which includes the activating complex <math>AR_{2}</math> | ||
| + | |||
| + | <sup>*2</sup> Reactions taking place only in the scenarios which includes the antisense RNA interactions | ||
| + | |||
| + | <sup>*3</sup> Reactions taking place only in the scenarios which include the activation of ''scbA'' by <math>R_{2}</math> | ||
| + | |||
| + | {|class="wikitable" | ||
| + | <table border='1' style="text-align:center"> | ||
| + | ! Reactions | ||
| + | |- | ||
| + | |<math>O_{R} + R_{2} \rightleftharpoons O_{R} \bullet R_{2}</math> | ||
| + | |||
| + | <math>O_{R}\bullet R_{2} + R_{2} \rightleftharpoons O_{R}\bullet 2R_{2}</math> | ||
| + | |||
| + | <math>O_{A} + R_{2} \rightleftharpoons O_{A}\bullet R_{2}</math> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2} + R_{2} \rightleftharpoons O_{A}\bullet 2R_{2}</math> | ||
| + | |||
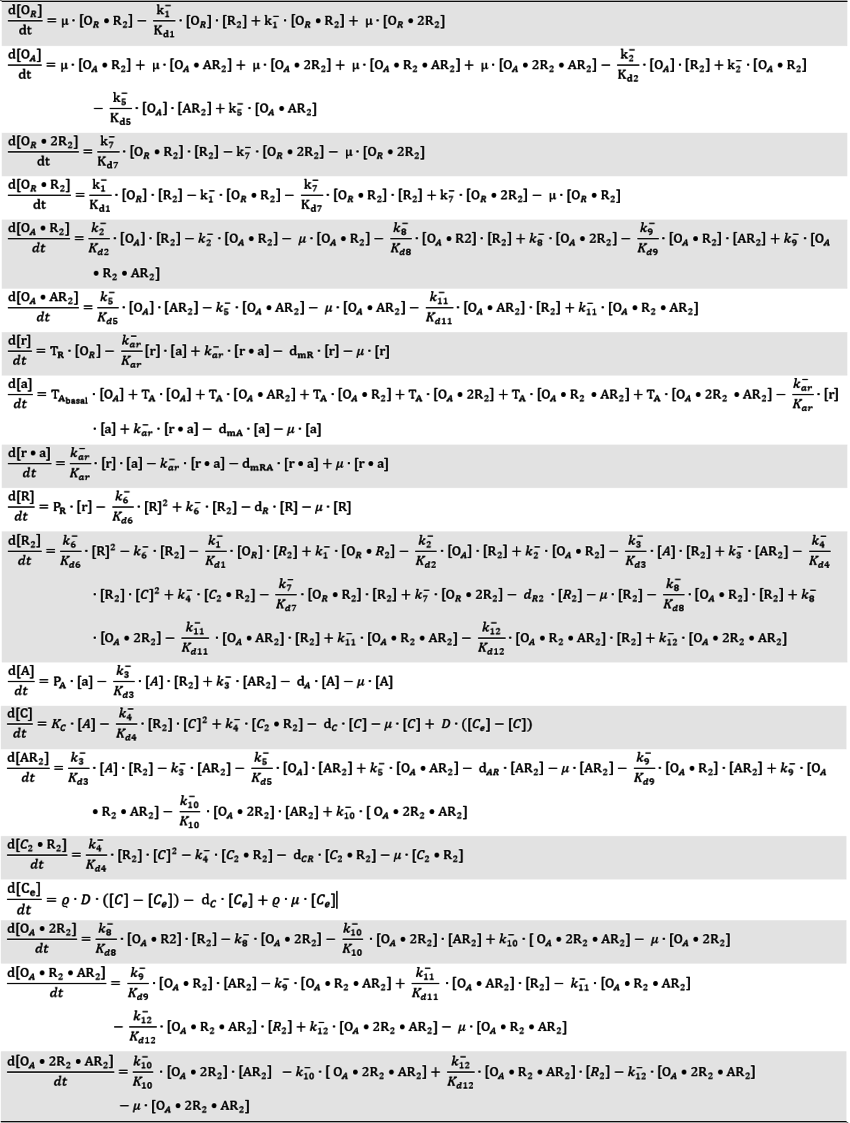
| + | <math>A + R_{2} \rightleftharpoons AR_{2}</math> <sup>*1</sup> | ||
| + | |||
| + | <math>2C + R_{2} \rightleftharpoons C_{2}\bullet R_{2}</math> | ||
| + | |||
| + | <math>O_{A} + AR_{2} \rightleftharpoons O_{A}\bullet AR_{2}</math> <sup>*1</sup> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2} + AR_{2} \rightleftharpoons O_{A}\bullet R_{2}\bullet AR_{2}</math> <sup>*1</sup> | ||
| + | |||
| + | <math>O_{A}\bullet 2R_{2} + AR_{2} \rightleftharpoons O_{A}\bullet 2R_{2}\bullet AR_{2}</math> <sup>*1</sup> | ||
| + | |||
| + | <math>O_{A}\bullet AR_{2} + R_{2} \rightleftharpoons O_{A}\bullet R_{2}\bullet AR_{2}</math> <sup>*1</sup> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2}\bullet AR_{2} + R_{2} \rightleftharpoons O_{A}\bullet 2R_{2}\bullet AR_{2}</math> <sup>*1</sup> | ||
| + | |||
| + | <math>S \xrightarrow{A} C</math> | ||
| + | |||
| + | <math>O_{R} \rightarrow O_{R} + r</math> | ||
| + | |||
| + | <math>O_{A} \rightarrow O_{A} + a</math> | ||
| + | |||
| + | <math>O_{A}\bullet AR_{2} \rightarrow O_{A}\bullet AR_{2} + a</math> <sup>*1</sup> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2} \rightarrow O_{A}\bullet R_{2} + a</math> <sup>*3</sup> | ||
| + | |||
| + | <math>O_{A}\bullet 2R_{2} \rightarrow O_{A}\bullet 2R_{2} + a</math> <sup>*3</sup> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2}\bullet AR_{2} \rightarrow O_{A}\bullet R_{2}\bullet AR_{2} + a</math> <sup>*3</sup> | ||
| + | |||
| + | <math>O_{A}\bullet 2R_{2}\bullet AR_{2} \rightarrow O_{A}\bullet 2R_{2}\bullet AR_{2} + a</math> <sup>*3</sup> | ||
| + | |||
| + | <math>a + r \rightleftharpoons r\bullet a</math> <sup>*2</sup> | ||
| + | |||
| + | <math>r \rightarrow r + R</math> | ||
| + | |||
| + | <math>a \rightarrow a + A</math> | ||
| + | |||
| + | <math>2R \rightleftharpoons R_{2}</math> | ||
| + | |||
| + | <math>C \underset{r \cdot D}{\stackrel{D}{\rightleftharpoons}} C_{e}</math> | ||
| + | |||
| + | <math>r\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>R\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>R_{2}\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>a\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>A\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>C\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>C_{2}\bullet R_{2}\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>AR_{2}\rightarrow \varnothing</math> <sup>*1</sup> | ||
| + | |||
| + | <math>r\bullet a\rightarrow \varnothing</math> <sup>*2</sup> | ||
| + | |||
| + | <math>C_{e}\rightarrow \varnothing</math> | ||
| + | |||
| + | <math>O_{R}\bullet R_{2} \rightarrow O_{R}\bullet R_{2} + O_{R}</math> | ||
| + | |||
| + | <math>O_{R}\bullet 2R_{2} \rightarrow O_{R}\bullet 2R_{2} + O_{R}</math> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2} \rightarrow O_{A}\bullet R_{2} + O_{A}</math> | ||
| + | |||
| + | <math>O_{A}\bullet 2R_{2} \rightarrow O_{A}\bullet 2R_{2} + O_{A}</math> | ||
| + | |||
| + | <math>O_{A}\bullet AR_{2} \rightarrow O_{A}\bullet AR_{2} + O_{A}</math> | ||
| + | |||
| + | <math>O_{A}\bullet R_{2}\bullet AR_{2} \rightarrow O_{A}\bullet R_{2}\bullet AR_{2} + O_{A}</math> | ||
| + | |||
| + | <math>O_{A}\bullet 2R_{2}\bullet AR_{2} \rightarrow O_{A}\bullet 2R_{2}\bullet AR_{2} + O_{A}</math> | ||
| + | |||
| + | |} | ||
== Parameters for model reactions == | == Parameters for model reactions == | ||
| Line 10: | Line 110: | ||
! Reaction | ! Reaction | ||
! Parameter | ! Parameter | ||
| − | ! | + | ! Values in literature |
! Units | ! Units | ||
| − | ! | + | ! Mode |
| − | Mode | + | ! Spread |
| − | ! | ||
| − | |||
! Location | ! Location | ||
parameter (μ) | parameter (μ) | ||
| Line 22: | Line 120: | ||
|- | |- | ||
|rowspan="2" |[[Binding of R2 to OR operator |Binding of R<sub>2</sub> to OR operator]] | |rowspan="2" |[[Binding of R2 to OR operator |Binding of R<sub>2</sub> to OR operator]] | ||
| − | |<math>K_{d1}</math> | + | |<math>K_{d1},K_{d7}</math> |
|0.005−5.8 | |0.005−5.8 | ||
|<math> nM </math> | |<math> nM </math> | ||
| − | |0. | + | |0.299 |
| − | | | + | |6.8 |
| − | | | + | |0.15471 |
| − | |1. | + | |1.1661 |
|- | |- | ||
| − | |<math>k^{-}_{1}</math> | + | |<math>k^{-}_{1}, k^{-}_{7}</math> |
| − | |0. | + | |0.09−0.846 |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.489 |
| − | | | + | |1.98 |
| − | |−0. | + | |−0.37642 |
| − | |0. | + | |0.58225 |
|- | |- | ||
|rowspan="2" |[[Binding of R2 to OA operator |Binding of R<sub>2</sub> to OA operator]] | |rowspan="2" |[[Binding of R2 to OA operator |Binding of R<sub>2</sub> to OA operator]] | ||
| − | |<math>K_{d2}</math> | + | |<math>K_{d2},K_{d8}</math> |
|0.005−5.8 | |0.005−5.8 | ||
|<math> nM </math> | |<math> nM </math> | ||
| − | |0. | + | |0.299 |
| − | | | + | |6.8 |
| − | | | + | |0.15471 |
| − | |1. | + | |1.1661 |
|- | |- | ||
| − | |<math>k^{-}_{2}</math> | + | |<math>k^{-}_{2},k^{-}_{8}</math> |
| − | |0. | + | |0.09−0.846 |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.489 |
| − | | | + | |1.98 |
| − | |−0. | + | |−0.37642 |
| − | |0. | + | |0.58225 |
|- | |- | ||
|rowspan="2" |[[Binding of R2 to A |Binding of R<sub>2</sub> to A]] | |rowspan="2" |[[Binding of R2 to A |Binding of R<sub>2</sub> to A]] | ||
|<math>K_{d3}</math> | |<math>K_{d3}</math> | ||
| − | |<math>10^{ | + | |<math>10^{4}−10^{6}</math> |
|<math> nM </math> | |<math> nM </math> | ||
| − | |<math> | + | |<math>9.9 \cdot 10^{4}</math> |
| − | | | + | |10.02 |
| − | | | + | |13.192 |
| − | | | + | |1.2977 |
|- | |- | ||
|<math>k^{-}_{3}</math> | |<math>k^{-}_{3}</math> | ||
| − | |<math> | + | |<math>38.3-4.46 \cdot 10^{6}</math> |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | | | + | |1879.7 |
| − | | | + | |23.13 |
| − | | | + | |9.9423 |
| − | |1. | + | |1.5503 |
|- | |- | ||
|rowspan="2" |[[Binding of R2 to C |Binding of R<sub>2</sub> to C]] | |rowspan="2" |[[Binding of R2 to C |Binding of R<sub>2</sub> to C]] | ||
|<math>K_{d4}</math> | |<math>K_{d4}</math> | ||
| − | |<math> | + | |<math>0.0011-10400</math> |
|<math> nM </math> | |<math> nM </math> | ||
| − | | | + | |1.58 |
| − | | | + | |730.63 |
| − | | | + | |5.9389 |
| − | | | + | |2.3412 |
|- | |- | ||
|<math>k^{-}_{4}</math> | |<math>k^{-}_{4}</math> | ||
|<math>3 \cdot 10^{-4}-126</math> | |<math>3 \cdot 10^{-4}-126</math> | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | | | + | |0.816 |
| − | | | + | |244 |
| − | | | + | |4.2872 |
| − | | | + | |2.119 |
|- | |- | ||
|rowspan="2" |[[Binding of A-R2 to OA' operator |Binding of A-R<sub>2</sub> to OA' operator]] | |rowspan="2" |[[Binding of A-R2 to OA' operator |Binding of A-R<sub>2</sub> to OA' operator]] | ||
| − | |<math>K_{d5}</math> | + | |<math>K_{d5},K_{d9},K_{d10},K_{d11},K_{d12}</math> |
|0.005−5.8 | |0.005−5.8 | ||
|<math> nM </math> | |<math> nM </math> | ||
| − | |0. | + | |0.299 |
| − | | | + | |6.8 |
| − | | | + | |0.15471 |
| − | |1. | + | |1.1661 |
|- | |- | ||
| − | |<math>k^{-}_{5}</math> | + | |<math>k^{-}_{5},k^{-}_{9},k^{-}_{10},k^{-}_{11},k^{-}_{12}</math> |
| − | |0. | + | |0.09−0.846 |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.489 |
| − | | | + | |1.98 |
| − | |−0. | + | |−0.37642 |
| − | |0. | + | |0.58225 |
|- | |- | ||
|[[Synthesis of C]] | |[[Synthesis of C]] | ||
|<math>K_{C}</math> | |<math>K_{C}</math> | ||
| − | |0. | + | |0.00315−85.5 |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.094 |
| − | | | + | |60.49 |
| − | | | + | |0.87914 |
| − | | | + | |1.8017 |
|- | |- | ||
| − | |[[Transcription of r]] | + | |rowspan="3" |[[Transcription of r]] |
| − | |<math> | + | |<math>\Omega_{R}</math> |
|0.16−8.24 | |0.16−8.24 | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.8346 |
| − | | | + | |3.55 |
| − | |0. | + | |0.63056 |
| − | |0. | + | |0.90076 |
| + | |- | ||
| + | |<math>k_{F}</math> | ||
| + | |18.2−33 | ||
| + | |<math> min^{-1} </math> | ||
| + | |20.7 | ||
| + | |1.34 | ||
| + | |3.1107 | ||
| + | |0.28276 | ||
| + | |- | ||
| + | |<math>k_{onR}</math> | ||
| + | |0.0518-14.5 | ||
| + | |<math> min^{-1} </math> | ||
| + | |0.867 | ||
| + | |4.09 | ||
| + | |0.78694 | ||
| + | |0.96428 | ||
|- | |- | ||
| − | |[[Transcription of a]] | + | |rowspan="3" |[[Transcription of a]] |
| − | |<math> | + | |<math>\Omega_{A_{basal}}</math> |
| + | |0.001−0.2 | ||
| + | |<math> min^{-1} </math> | ||
| + | |0.0037 | ||
| + | |10.93 | ||
| + | |−3.8324 | ||
| + | |1.3259 | ||
| + | |- | ||
| + | |<math>\Omega_{A}</math> | ||
|0.11−5.65 | |0.11−5.65 | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.572 |
| − | | | + | |3.5 |
| − | |0. | + | |0.2514 |
| − | |0. | + | |0.89956 |
| + | |- | ||
| + | |<math>k_{onA}</math> | ||
| + | |0.039-8.82 | ||
| + | |<math> min^{-1} </math> | ||
| + | |0.587 | ||
| + | |3.875 | ||
| + | |0.35255 | ||
| + | |0.94054 | ||
|- | |- | ||
|rowspan="2" |[[Antisense interaction between r and a]] | |rowspan="2" |[[Antisense interaction between r and a]] | ||
|<math>K_{ar}</math> | |<math>K_{ar}</math> | ||
| − | |0. | + | |0.4−89 |
| − | |<math> | + | |<math> nM </math> |
| − | | | + | |7.8 |
| − | | | + | |7.27 |
| − | | | + | |3.4665 |
| − | | | + | |1.1882 |
|- | |- | ||
|<math>k^{-}_{ar}</math> | |<math>k^{-}_{ar}</math> | ||
|0.006−6 | |0.006−6 | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.223 |
| − | | | + | |4.8 |
| − | | | + | |−0.43359 |
| − | | | + | |1.0326 |
|- | |- | ||
|[[Translation of R]] | |[[Translation of R]] | ||
| Line 154: | Line 284: | ||
|0.165−5.86 | |0.165−5.86 | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.744 |
| − | | | + | |3.2 |
| − | |0. | + | |0.42952 |
| − | |0. | + | |0.85176 |
|- | |- | ||
|[[Translation of A]] | |[[Translation of A]] | ||
| Line 163: | Line 293: | ||
|0.11−4 | |0.11−4 | ||
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | |0. | + | |0.5 |
| − | | | + | |3.2 |
| − | |0. | + | |0.053356 |
| − | |0. | + | |0.85327 |
|- | |- | ||
|rowspan="2" |[[Formation of homo-dimer R2 |Formation of homo-dimer R<sub>2</sub>]] | |rowspan="2" |[[Formation of homo-dimer R2 |Formation of homo-dimer R<sub>2</sub>]] | ||
|<math>K_{d6}</math> | |<math>K_{d6}</math> | ||
| − | | | + | |2.58-42.9 |
|<math> nM </math> | |<math> nM </math> | ||
| − | | | + | |3.89 |
| − | | | + | |1.9 |
| − | | | + | |1.6716 |
| − | | | + | |0.55779 |
|- | |- | ||
|<math>k^{-}_{6}</math> | |<math>k^{-}_{6}</math> | ||
| − | | | + | |0.144-51.2 |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | | | + | |1.1997 |
| − | | | + | |21.7 |
| − | |2. | + | |2.5303 |
| − | |1. | + | |1.5324 |
|- | |- | ||
|[[Degradation of r]] | |[[Degradation of r]] | ||
|<math>d_{mR}</math> | |<math>d_{mR}</math> | ||
| − | |0.03−0. | + | |0.03−0.365 |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |0. | + | |0.14 |
| − | | | + | |2.06 |
| − | |−1. | + | |−1.5916 |
| − | |0. | + | |0.60824 |
|- | |- | ||
|[[Degradation of R]] | |[[Degradation of R]] | ||
|<math>d_{R}</math> | |<math>d_{R}</math> | ||
| − | |<math> | + | |<math>5.02 \cdot 10^{-4}- 0.00578</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.00144</math> |
| − | | | + | |1.78 |
| − | | | + | |−6.2837 |
| − | | | + | |0.50981 |
|- | |- | ||
|[[Degradation of R2 |Degradation of R<sub>2</sub>]] | |[[Degradation of R2 |Degradation of R<sub>2</sub>]] | ||
|<math>d_{R_{2}}</math> | |<math>d_{R_{2}}</math> | ||
| − | |<math> | + | |<math>5.02 \cdot 10^{-4}- 0.00578</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.00144</math> |
| − | | | + | |1.78 |
| − | | | + | |−6.2837 |
| − | | | + | |0.50981 |
|- | |- | ||
|[[Degradation of a]] | |[[Degradation of a]] | ||
|<math>d_{mA}</math> | |<math>d_{mA}</math> | ||
| − | |0.03−0. | + | |0.03−0.365 |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |0. | + | |0.14 |
| − | | | + | |2.06 |
| − | |−1. | + | |−1.5916 |
| − | |0. | + | |0.60824 |
|- | |- | ||
|[[Degradation of A]] | |[[Degradation of A]] | ||
|<math>d_{A}</math> | |<math>d_{A}</math> | ||
| − | |<math> | + | |<math>5.02 \cdot 10^{-4}- 0.00578</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.00144</math> |
| − | | | + | |1.78 |
| − | | | + | |−6.2837 |
| − | | | + | |0.50981 |
|- | |- | ||
|[[Degradation of C]] | |[[Degradation of C]] | ||
|<math>d_{C}</math> | |<math>d_{C}</math> | ||
| − | |<math> | + | |<math>7.68 \cdot 10^{-5}- 2 \cdot 10^{-2}</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.0032</math> |
| − | | | + | |5.1 |
| − | | | + | |−4.6234 |
| − | | | + | |1.0539 |
|- | |- | ||
|[[Degradation of C2-R2 |Degradation of C<sub>2</sub>-R<sub>2</sub>]] | |[[Degradation of C2-R2 |Degradation of C<sub>2</sub>-R<sub>2</sub>]] | ||
|<math>d_{CR}</math> | |<math>d_{CR}</math> | ||
| − | |<math> | + | |<math>5.02 \cdot 10^{-4}- 0.00578</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.00144</math> |
| − | | | + | |1.78 |
| − | | | + | |−6.2837 |
| − | | | + | |0.50981 |
|- | |- | ||
|[[Degradation of A-R2 |Degradation of A-R<sub>2</sub>]] | |[[Degradation of A-R2 |Degradation of A-R<sub>2</sub>]] | ||
|<math>d_{AR}</math> | |<math>d_{AR}</math> | ||
| − | |<math> | + | |<math>5.02 \cdot 10^{-4}- 0.00578</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.00144</math> |
| − | | | + | |1.78 |
| − | | | + | |−6.2837 |
| − | | | + | |0.50981 |
|- | |- | ||
|[[Degradation of r-a]] | |[[Degradation of r-a]] | ||
|<math>d_{mRA}</math> | |<math>d_{mRA}</math> | ||
| − | |0. | + | |0.35−69.3 |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | | | + | |4.9 |
| − | | | + | |14.1 |
| − | |2. | + | |2.5669 |
| − | |0. | + | |0.98835 |
|- | |- | ||
|[[Diffusion of C and Ce |Diffusion of C and C<sub>e</sub>]] | |[[Diffusion of C and Ce |Diffusion of C and C<sub>e</sub>]] | ||
| Line 270: | Line 400: | ||
|0.1−24 | |0.1−24 | ||
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |8 | + | |2.8 |
| − | | | + | |4.27 |
| − | | | + | |1.9947 |
| − | |0. | + | |0.98233 |
|- | |- | ||
|[[Degradation of Ce]] | |[[Degradation of Ce]] | ||
| − | |<math>d_{C}</math> | + | |<math>d_{C}</math> |
| − | |<math> | + | |<math>7.68 \cdot 10^{-5}- 2 \cdot 10^{-2}</math> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| − | |<math> | + | |<math>0.0032</math> |
| − | + | |5.1 | |
| − | + | |−4.6234 | |
| − | + | |1.0539 | |
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | |||
| − | | | ||
| − | | | ||
| − | |||
| − | |||
|} | |} | ||
| − | == Parameters for cell | + | == Parameters for cell growth == |
{|class="wikitable" | {|class="wikitable" | ||
<table border='1' style="text-align:center"> | <table border='1' style="text-align:center"> | ||
! Parameter | ! Parameter | ||
| − | ! | + | ! Range of values |
! Units | ! Units | ||
|- | |- | ||
| − | | <math> | + | |<math>K</math> |
| − | |<math> | + | |<math>7.14 \cdot 10^{12} - 9\cdot 10^{12}</math> |
| − | | | + | |cells |
|- | |- | ||
| − | | <math> | + | |<math>N_0</math> |
| − | |<math> | + | |<math>5 \cdot 10^{10} - 2\cdot 10^{11}</math> |
| − | | | + | |cells |
|- | |- | ||
| − | | <math> | + | |<math>\mu_{max}</math> |
| − | |<math> | + | |<math>0.003368 - 0.007499</math> |
| − | |<math> | + | |<math>min^{-1}</math> |
|- | |- | ||
| − | |<math> | + | |<math>v</math> |
| − | |<math> 2 \ | + | |<math>0.001482 - 0.5814</math> |
| − | |<math> | + | |<math>min^{-1}</math> |
| + | |- | ||
| + | |<math>m</math> | ||
| + | |<math>0.47 - 2.46</math> | ||
| + | |n.a. | ||
| + | |- | ||
| + | |<math>\lambda</math> | ||
| + | |<math>330.52 - 883</math> | ||
| + | |<math>min</math> | ||
|} | |} | ||
| Line 329: | Line 458: | ||
|- | |- | ||
|O<sub>R</sub> | |O<sub>R</sub> | ||
| − | | | + | |0.9592 |
|<math> nM </math> | |<math> nM </math> | ||
|Cell | |Cell | ||
|- | |- | ||
| O<sub>A</sub> | | O<sub>A</sub> | ||
| − | | | + | |0.9592 |
|<math> nM </math> | |<math> nM </math> | ||
|Cell | |Cell | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| O<sub>R</sub>-R<sub>2</sub> | | O<sub>R</sub>-R<sub>2</sub> | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| O<sub>A</sub>-R<sub>2</sub> | | O<sub>A</sub>-R<sub>2</sub> | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| O<sub>A</sub>'-AR<sub>2</sub> | | O<sub>A</sub>'-AR<sub>2</sub> | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| r | | r | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| a | | a | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| r-a | | r-a | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| R | | R | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| R<sub>2</sub> | | R<sub>2</sub> | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| A | | A | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| C | | C | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| AR<sub>2</sub> | | AR<sub>2</sub> | ||
| − | | | + | |0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| C<sub>2</sub>R<sub>2</sub> | | C<sub>2</sub>R<sub>2</sub> | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Cell | | Cell | ||
|- | |- | ||
| C<sub>e</sub> | | C<sub>e</sub> | ||
| − | | | + | | 0 |
|<math> nM </math> | |<math> nM </math> | ||
| Environment | | Environment | ||
|} | |} | ||
| − | |||
== Differential equations == | == Differential equations == | ||
| − | + | [[Image:Diffeq.png|850px]] | |
Latest revision as of 17:10, 16 October 2019
Contents
Model scripts
The model scripts for the different scenarios can be found in pdf form here: File:GBL Models.pdf
List of Model Reactions
*1 Reactions taking place only in the scenarios which includes the activating complex 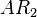
*2 Reactions taking place only in the scenarios which includes the antisense RNA interactions
*3 Reactions taking place only in the scenarios which include the activation of scbA by 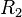
| Reactions |
|---|
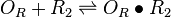
|
Parameters for model reactions
| Reaction | Parameter | Values in literature | Units | Mode | Spread | Location
parameter (μ) |
Scale
parameter (σ) |
|---|---|---|---|---|---|---|---|
| Binding of R2 to OR operator | 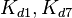
|
0.005−5.8 | 
|
0.299 | 6.8 | 0.15471 | 1.1661 |
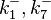
|
0.09−0.846 | 
|
0.489 | 1.98 | −0.37642 | 0.58225 | |
| Binding of R2 to OA operator | 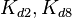
|
0.005−5.8 | 
|
0.299 | 6.8 | 0.15471 | 1.1661 |
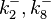
|
0.09−0.846 | 
|
0.489 | 1.98 | −0.37642 | 0.58225 | |
| Binding of R2 to A | 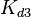
|
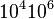
|

|
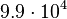
|
10.02 | 13.192 | 1.2977 |

|
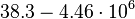
|

|
1879.7 | 23.13 | 9.9423 | 1.5503 | |
| Binding of R2 to C | 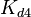
|
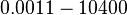
|

|
1.58 | 730.63 | 5.9389 | 2.3412 |
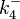
|
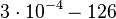
|

|
0.816 | 244 | 4.2872 | 2.119 | |
| Binding of A-R2 to OA' operator | 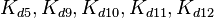
|
0.005−5.8 | 
|
0.299 | 6.8 | 0.15471 | 1.1661 |
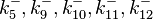
|
0.09−0.846 | 
|
0.489 | 1.98 | −0.37642 | 0.58225 | |
| Synthesis of C | 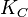
|
0.00315−85.5 | 
|
0.094 | 60.49 | 0.87914 | 1.8017 |
| Transcription of r | 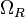
|
0.16−8.24 | 
|
0.8346 | 3.55 | 0.63056 | 0.90076 |
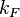
|
18.2−33 | 
|
20.7 | 1.34 | 3.1107 | 0.28276 | |
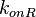
|
0.0518-14.5 | 
|
0.867 | 4.09 | 0.78694 | 0.96428 | |
| Transcription of a | 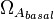
|
0.001−0.2 | 
|
0.0037 | 10.93 | −3.8324 | 1.3259 |
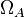
|
0.11−5.65 | 
|
0.572 | 3.5 | 0.2514 | 0.89956 | |
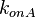
|
0.039-8.82 | 
|
0.587 | 3.875 | 0.35255 | 0.94054 | |
| Antisense interaction between r and a | 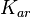
|
0.4−89 | 
|
7.8 | 7.27 | 3.4665 | 1.1882 |
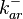
|
0.006−6 | 
|
0.223 | 4.8 | −0.43359 | 1.0326 | |
| Translation of R | 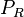
|
0.165−5.86 | 
|
0.744 | 3.2 | 0.42952 | 0.85176 |
| Translation of A | 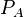
|
0.11−4 | 
|
0.5 | 3.2 | 0.053356 | 0.85327 |
| Formation of homo-dimer R2 | 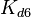
|
2.58-42.9 | 
|
3.89 | 1.9 | 1.6716 | 0.55779 |
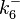
|
0.144-51.2 | 
|
1.1997 | 21.7 | 2.5303 | 1.5324 | |
| Degradation of r | 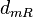
|
0.03−0.365 | 
|
0.14 | 2.06 | −1.5916 | 0.60824 |
| Degradation of R | 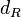
|
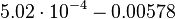
|

|
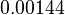
|
1.78 | −6.2837 | 0.50981 |
| Degradation of R2 | 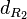
|
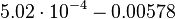
|

|
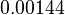
|
1.78 | −6.2837 | 0.50981 |
| Degradation of a | 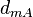
|
0.03−0.365 | 
|
0.14 | 2.06 | −1.5916 | 0.60824 |
| Degradation of A | 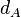
|
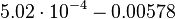
|

|
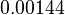
|
1.78 | −6.2837 | 0.50981 |
| Degradation of C | 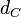
|
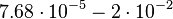
|

|
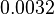
|
5.1 | −4.6234 | 1.0539 |
| Degradation of C2-R2 | 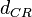
|
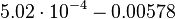
|

|
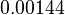
|
1.78 | −6.2837 | 0.50981 |
| Degradation of A-R2 | 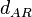
|
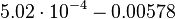
|

|
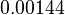
|
1.78 | −6.2837 | 0.50981 |
| Degradation of r-a | 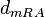
|
0.35−69.3 | 
|
4.9 | 14.1 | 2.5669 | 0.98835 |
| Diffusion of C and Ce | 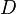
|
0.1−24 | 
|
2.8 | 4.27 | 1.9947 | 0.98233 |
| Degradation of Ce | 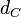
|
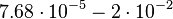
|

|
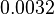
|
5.1 | −4.6234 | 1.0539 |


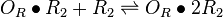
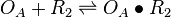
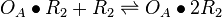
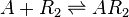 *1
*1
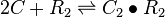
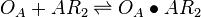 *1
*1
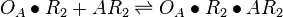 *1
*1
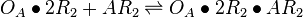 *1
*1
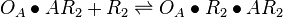 *1
*1
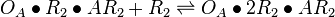 *1
*1
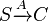
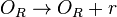
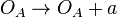
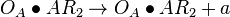 *1
*1
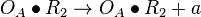 *3
*3
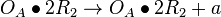 *3
*3
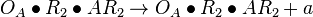 *3
*3
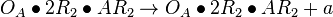 *3
*3
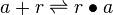 *2
*2
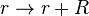
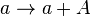
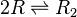
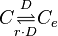
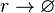
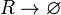
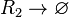
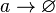
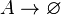
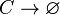
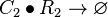
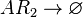 *1
*1
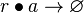 *2
*2