Difference between revisions of "Extracellular signal-regulated kinase (ERK)"
m |
|||
| Line 5: | Line 5: | ||
<h2> Description of the model </h2> | <h2> Description of the model </h2> | ||
<!--[[File:L MAPK SignallingNetwork.png]]--> | <!--[[File:L MAPK SignallingNetwork.png]]--> | ||
| + | <div align="left"> | ||
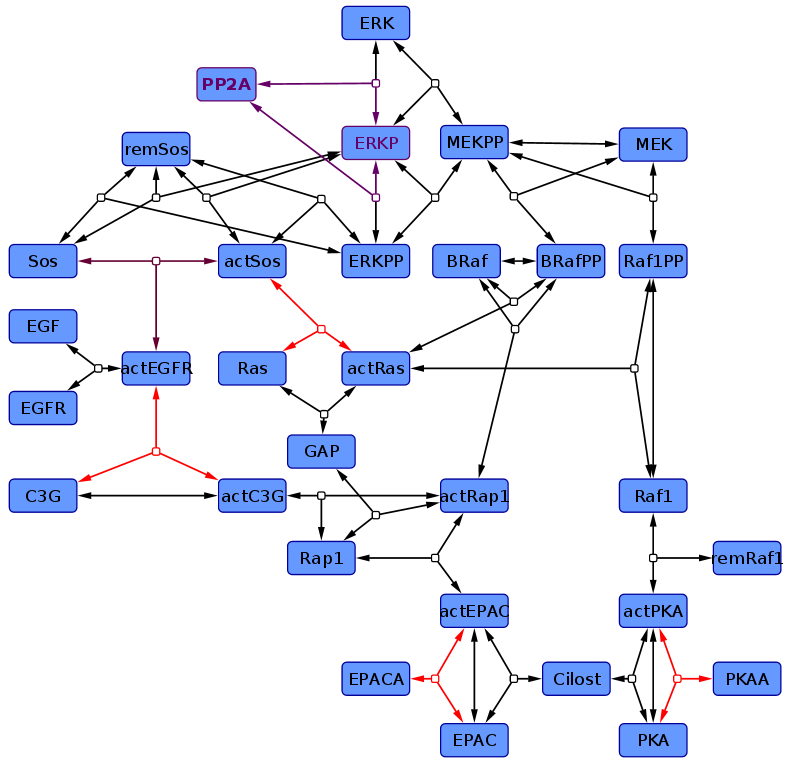
<p> According to the topology which is found as most likely by Xu et al (2010)<ref name="Xu2010"> Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. ([http://www.ncbi.nlm.nih.gov/pubmed/20234003 pmid:20234003])</ref> a kinetic model is created. Literature data is used to estimate the parameter values and their error. We ended up in two different models. In one (non-modified model) only one reaction kinetic differs from Xu et al (2010)<ref name="Xu2010"></ref>, the Sos-Activation (purple reaction) and is assumed be a binding process rather than enzyme catalysed. Furthermore we modelled the ERK-Activation as two step reaction, with the monophosphorylated ERK as intermediate.<br> | <p> According to the topology which is found as most likely by Xu et al (2010)<ref name="Xu2010"> Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. ([http://www.ncbi.nlm.nih.gov/pubmed/20234003 pmid:20234003])</ref> a kinetic model is created. Literature data is used to estimate the parameter values and their error. We ended up in two different models. In one (non-modified model) only one reaction kinetic differs from Xu et al (2010)<ref name="Xu2010"></ref>, the Sos-Activation (purple reaction) and is assumed be a binding process rather than enzyme catalysed. Furthermore we modelled the ERK-Activation as two step reaction, with the monophosphorylated ERK as intermediate.<br> | ||
In the other one (modified model) further kinetics were changed in addition to these modifications. So are the activation of EPAC and PKA by an agonist modelled as binding, C3G-Activation is modelled in two steps, first a binding to the receptor than a phosphorylation, and the Ras-Activation process is splitted in its elementary reactions, and their kinetics are therefore mass action. <br> | In the other one (modified model) further kinetics were changed in addition to these modifications. So are the activation of EPAC and PKA by an agonist modelled as binding, C3G-Activation is modelled in two steps, first a binding to the receptor than a phosphorylation, and the Ras-Activation process is splitted in its elementary reactions, and their kinetics are therefore mass action. <br> | ||
The differentiation in two different models is made to gain one model which is as close as possible to Xu et al. (2010)<ref name="Xu2010"> Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. ([http://www.ncbi.nlm.nih.gov/pubmed/20234003 pmid:20234003])</ref> and another one, in which the reaction mechanism resemble the published one and therefore more measured parameter are available. | The differentiation in two different models is made to gain one model which is as close as possible to Xu et al. (2010)<ref name="Xu2010"> Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. ([http://www.ncbi.nlm.nih.gov/pubmed/20234003 pmid:20234003])</ref> and another one, in which the reaction mechanism resemble the published one and therefore more measured parameter are available. | ||
</p> | </p> | ||
| + | </div> | ||
| − | <div align="right"> | + | <div align="right" width="10%"> |
<imagemap> | <imagemap> | ||
| − | File:L_MAPK_SignallingNetwork.png | + | File:L_MAPK_SignallingNetwork.png| |
rect 130 435 179 467 [[C3G-Activation|C3G-Activation]] | rect 130 435 179 467 [[C3G-Activation|C3G-Activation]] | ||
Revision as of 17:12, 11 March 2014
Description of the model
According to the topology which is found as most likely by Xu et al (2010)[1] a kinetic model is created. Literature data is used to estimate the parameter values and their error. We ended up in two different models. In one (non-modified model) only one reaction kinetic differs from Xu et al (2010)[1], the Sos-Activation (purple reaction) and is assumed be a binding process rather than enzyme catalysed. Furthermore we modelled the ERK-Activation as two step reaction, with the monophosphorylated ERK as intermediate.
In the other one (modified model) further kinetics were changed in addition to these modifications. So are the activation of EPAC and PKA by an agonist modelled as binding, C3G-Activation is modelled in two steps, first a binding to the receptor than a phosphorylation, and the Ras-Activation process is splitted in its elementary reactions, and their kinetics are therefore mass action.
The differentiation in two different models is made to gain one model which is as close as possible to Xu et al. (2010)[1] and another one, in which the reaction mechanism resemble the published one and therefore more measured parameter are available.
Reactions
Model File
References
- ↑ 1.0 1.1 1.2 Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. (pmid:20234003)