EPAC and PKA-Activation(Cilostamide)
Binding Reaction
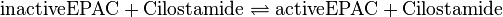
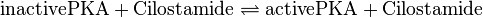
Kinetic equation
![v_{\text{EPAC-Activation}} = \frac{k_{cat} \cdot [\text{Cilostamide}] \cdot [\text{inactiveEPAC}] \cdot (1-\frac{[\text{activeEPAC}]}{[\text{inactiveEPAC}] \cdot K_{eq}})}{Km_{\text{inactiveEPAC}} \cdot (1 + \frac{[\text{inactiveEPAC}]}{Km_{\text{inactiveEPAC}}} + \frac{[\text{activeEPAC}]}{Km_{\text{activeEPAC}}})}](/wiki/images/math/7/5/4/75468df500f444951f3c320ba1c3f71a.png)
![v_{\text{PKA-Activation}} = \frac{k_{cat} \cdot [\text{Cilostamide}] \cdot [\text{inactivePKA}] \cdot (1-\frac{[\text{activePKA}]}{[\text{inactivePKA}] \cdot K_{eq}})}{Km_{\text{inactivePKA}} \cdot (1 + \frac{[\text{inactivePKA}]}{Km_{\text{inactivePKA}}} + \frac{[\text{activePKA}]}{Km_{\text{activePKA}}})}](/wiki/images/math/b/1/9/b1949c3e0f223ea757316799d92d25ca.png)
final Parameter
KminactiveEPAC = 9699.87 M = 9.69987 * 109 μM
KminactivePKA = 3102.31 M = 3.10231 * 109 μM
kcatPKA = 1.6605 1/s
kcatEPAC = 9.34508 1/s
Annotations
Cilostamide is an inhibitor of phosphodiesterase 3 (PDE3). The function of PDE3 is the hydrolysation of cAMP and cGMP to AMP and GMP, respectively. It is expressed in various tissues.[1] So by inhibiting PDE3 the cAMP level increases, which leads to an activation of PKA and EPAC. The PDE3, however, is not included in the model and therefore the indirect activation process via PDE3 inhibition is modelled as a direct activation process. Due to this reduction, there are no parameters for this process measured. The parameters used by us are the parameters by Xu et al (2010)[2] and no standard deviation is assumed, because it is a fictive reaction for which assuming an error does not make sense. The same Michaelis-Menten constant for substrate and product is used.
There is no incident about the equilibrium constant. However because Xu et al. (2010) [2] has modelled this reaction as irreversible we can assume that the equilibrium is far on the product side and therefore the equilibrium constant is high. We used Keq = 109.
KmP value and equilibrium constant
The Km value for the product is assumed to be similar to but slightly higher than the the Km value of the substrate because of the similarity of the both species. Therefore the Km value of the substrate is multiplied by 1.05 to gain the one of the product.
For information about the equilibrium constant please see here.
References
- ↑ Francis S.H., Blount M.A. and Corbin J.D. (2011) Mammalian cyclic nucleotide phosphodiesterases: molecular mechanisms and physiological functions." Physiol Rev. 91.2,651-690. DOI:10.1152/physrev.00030.2010 (pmid:21527734)
- ↑ 2.0 2.1 Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. (pmid:20234003)