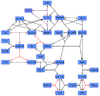
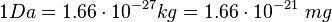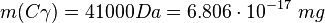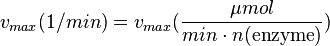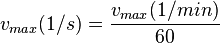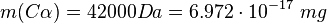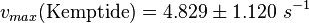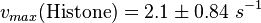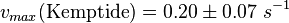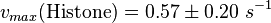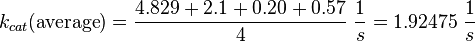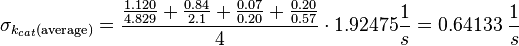Raf-1-Removal
Binding Reaction
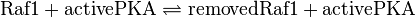
Kinetic Equation
![\frac{k_{cat} \cdot [\text{activePKA}] \cdot [\text{Raf1}] \cdot (1-\frac{[\text{removedRaf1}]}{[\text{Raf1}] \cdot K_{eq}})}{Km_{\text{Raf1}}\cdot(1 + \frac{[\text{Raf1}]}{Km_{\text{Raf1}}} + \frac{[\text{removedRaf1}]}{Km_{\text{removedRaf1}}})}](/wiki/images/math/c/8/0/c8091003c462439b89b0ea7f56a23bcf.png)
final Parameter
Because only a subunit was measured the standard deviation is increased by 1.5.
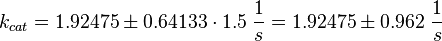
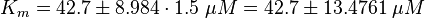
Parameter
| Calculation | Reference |
|---|---|
|
The calculation of the vmax values that have the right unity is as follows:
Failed to parse (lexing error): \text{#enzyme per mg} = \frac{1 mg}{m(C \gamma)} = 1.469 \cdot 10^{16} Failed to parse (lexing error): n(\text{enzyme per mg}) = \frac{\text{#enzyme/mg}}{6.023\cdot 10^{17}}= 0.024395\ \mu mol
Failed to parse (lexing error): \text{#enzyme per mg} = \frac{1 mg}{m(C \gamma)} = 1.4343 \cdot 10^{16} Failed to parse (lexing error): n(\text{enzyme per mg}) = \frac{\text{#enzyme/mg}}{6.023\cdot 10^{17}}= 0.023813\ \mu mol
This calculation results in: Cα
Cγ
|
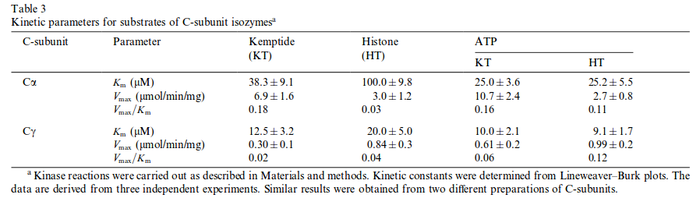 Zhang W. et al.(2004)[1] |
KmP value and equilibrium constant
The Km value for the product is assumed to be similar but slightly higher than the the Km value of the substrate because of the similarity of the both species. Therefore the Km value of the substrate is multiplied by 1.05 to gain the one of the product and the uncertainty is increased by increasing the error on Kmsubstrate by 50%.
For information about the equilibrium constant please see here.
References
- ↑ Zhang W., Morris G.Z., and Beebe S.J.(2004) "Characterization of the cAMP-dependent protein kinase catalytic subunit Cgamma expressed and purified from sf9 cells." Protein Expr Purif 35.1:156-169. DOI:10.1016/j.pep.2004.01.006. (pmid:15039070)