C3G-Activation
Binding Reaction
non-modified model:
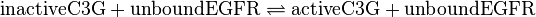 , catalysed by src kinase
, catalysed by src kinasemodified model:
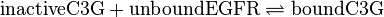
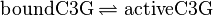 , catalysed by src kinase
, catalysed by src kinase Kinetic equation
non-modified model:
![v_{\text{C3G-Activation}}=\frac{k_{cat} \cdot [\text{unboundEGFR}] \cdot [\text{inactiveC3G}] \cdot (1-\frac{[\text{activeC3G}]}{[\text{inactiveC3G}] \cdot K_{eq}})}{Km_{\text{inactiveC3G}} \cdot (1 + \frac{[\text{inactiveC3G}]}{Km_{\text{inactiveC3G}}} + \frac{[\text{activeC3G}]}{Km_{\text{activeC3G}}})}](/wiki/images/math/c/8/4/c8412daeaec6fc4bdac1d8c1e45f085f.png)
modified model:
![v_{\text{C3G-Binding}}=k_{ass} \cdot [\text{inactiveC3G}] \cdot [\text{unboundEGFR}]-k_{diss} \cdot [\text{boundC3G}]](/wiki/images/math/6/9/7/697f68871026a241efbe71e3309f4f0e.png)
![v_{\text{C3G-Activation}}=\frac{v_{max} \cdot [\text{boundC3G}] \cdot (1-\frac{[\text{activeC3G}]}{K_{eq}})}{Km_{\text{boundC3G}} \cdot (1 + \frac{[\text{boundC3G}]}{Km_{\text{boundC3G}}} + \frac{[\text{activeC3G}]}{Km_{\text{activeC3G}}})}](/wiki/images/math/1/0/d/10d348bc872f05d839bb0eb3964a594d.png)
General Information
C3G is like Sos a GNRP. It binds to the N-terminal SH3 domain of Crk, an adaptor protein which has no catalytic domain, but 1 SH2 and 2 SH3 domains (Feller et al. 1995[1], York et al. 1998 [2]). Unlike Sos however C3G is not mainly activated due to the recruitment to the membrane but due to the phosphorylation by Src-kinases. The way of how C3G processes the input seems to be similar to Sos, because the C-terminus is homolog to Cdc25, the catalytic domain of Sos.(Ichiba et al. 1999)[3]) However the main substrate of C3G is not Ras, but Rap1.(York et al. 1998)[2]
This activation process is once modelled as single Michaelis-Menten reaction (non-modified model) and once as a two step reaction: first binding, then phosphorylation (modified model).
For the phosphorylation step data found at (BRENDA EC 2.7.10.2) is used because the src is the main kinase that activates C3G.
For the binding the same parameters as for the Sos-Binding process are used. (see here)
final Parameter
Binding Step:
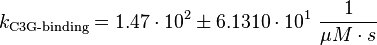
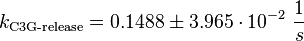
Phosphorylation Step:(used in the modified and non-modified model)
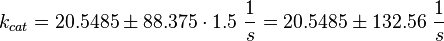
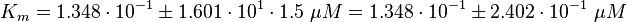
Parameter Calculation
The values that are used can be found in the screenshot below. All values except the ones in which ATP is used as substrate are averaged and the standard deviation is calculated.
BRENDA EC 2.7.10.2 - Screenshots
2.7.10.2 - non-specific protein-tyrosine kinase
KmP value and equilibrium constant
The Km value for the product is assumed to be similar to but slightly higher than the the Km value of the substrate because of the similarity of the both species. Therefore the Km value of the substrate is multiplied by 1.05 to gain the one of the product and the uncertainty is increased by increasing the error on Kmsubstrate by 50%.
For information about the equilibrium constant please see here.
References
- ↑ Feller S.M., Knudsen B., and Hanafusa H. (1995) "Cellular proteins binding to the first Src homology 3 (SH3) domain of the proto-oncogene product c-Crk indicate Crk-specific signaling pathways." Oncogene 10.8:1465-1473 (pmid:7731701)
- ↑ 2.0 2.1 York R.D. et al. (1998) "Rap1 mediates sustained MAP kinase activation induced by nerve growth factor."Nature 392.6676:622-626 (pmid:9560161)
- ↑ Ichiba T. et al. (1999) "Activation of C3G guanine nucleotide exchange factor for Rap1 by phosphorylation of tyrosine 504." J Biol Chem 274.20:14376-14381 (pmid:10318861