ERK-Activation
Reaction
Step 1:
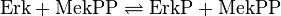
Step 2:
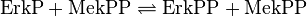
Kinetic Equation
Step 1:
![v_{\text{Erk-Activation}} = \frac{k_{cat} \cdot [\text{MekPP}] \cdot [\text{Erk}] \cdot (1-\frac{[\text{ErkP}]}{[\text{Erk}] \cdot K_{eq}})}{Km_{\text{Erk}} \cdot (1 + \frac{[\text{Erk}]}{Km_{\text{Erk}}} + \frac{[\text{ErkP}]}{Km_{\text{ErkP}}})}](/wiki/images/math/7/1/0/710929fc6024e3ad8fe374bc01babe15.png)
Step 2:
![v_{\text{Erk-Activation}} = \frac{k_{cat} \cdot [\text{MekPP}] \cdot [\text{ErkP}] \cdot (1-\frac{[\text{ErkPP}]}{[\text{ErkP}] \cdot K_{eq}})}{Km_{\text{ErkP}} \cdot (1 + \frac{[\text{ErkP}]}{Km_{\text{ErkP}}} + \frac{[\text{ErkPP}]}{Km_{\text{ErkPP}}})}](/wiki/images/math/4/6/7/46726f2681d7df1eb3432a57332471a2.png)
General Information
To be completely activated Erk has to be phoshporylated twice by Mek. First on a tyrosine residue and the on threonine Haystead T.A. et al. (1992)[1]. Therefore the Erk-Activation process is included in our model as a two step process and Erk is assumed to be present in thee different conformations: unphosphorylated, monophosphorylated and bisphosphorylated.
final Parameter
Failed to parse (lexing error): K_{\text{Mp42mapk_{unphopshorylated}}} = 476 \cdot 10^{-3} \pm 67.416 \cdot 10^{-3} \mu M
Failed to parse (lexing error): K_{\text{Mp42mapk{pY}}} = 46.6 \cdot 10^{-3} \pm 6.6 \cdot 10^{-3} \mu M (n=5)
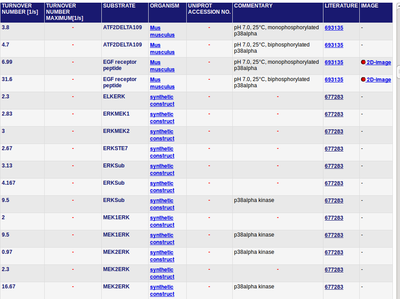
Data model Xu et al.[2]:
Failed to parse (lexing error): kcat_{\text{Mp42mapk_{unphopshorylated}}} = 6.48 \cdot 10^{-1} \pm 0.175541 \cdot 6.48 \cdot 10^{-1}\; \frac{1}{s} = 6.48 \cdot 10^{-1} \pm 1.1383\cdot 10^{-1}\; \frac{1}{s}
Failed to parse (lexing error): kcat_{\text{Mp42mapk{pY}}}=6.48 \cdot 10^{-1} \pm 0.175541 \cdot 6.48 \cdot 10^{-1}\; \frac{1}{s} = 6.48 \cdot 10^{-1} \pm 1.1383\cdot 10^{-1}\; \frac{1}{s}
Data BRENDA: Failed to parse (lexing error): kcat_{\text{Mp42mapk_{unphopshorylated}}} = 4.919 \pm 0.51\; \frac{1}{s}
Failed to parse (lexing error): kcat_{\text{Mp42mapk{pY}}} = 4.919 \pm 0.51\; \frac{1}{s}
Parameter
KMp42mapkunphopshorylated = 476 nM [1]
KMp42mapkpY = 46.6 ± 6.6 nM (n=5)[1]
Calculation of the uncertainty of KMp42mapkunphopshorylated
Because both Km values were measured be the same group and under the same conditions we assume that both values have the same relative error. The absolute error of KMp42mapkpY is given and the relative error is calculated to be 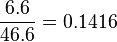 , which leads to an absolute error of KMp42mapkunphopshorylated.
, which leads to an absolute error of KMp42mapkunphopshorylated.
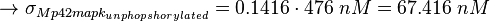
Firstly the kcat-values by Xu et al. Xu et al (2010)[2] were used. And for the uncertainty the averaged relative standard deviation is calculated is used. (see here)
However it was shown that this assumption leads to a too low kcat value. Both ERK and MEK are MAP kinases which might have a similar turnover number. There are no turnover numbers for MEK, so to finally estimate the catalytic constant data of the MAP kinase is used averaged and the standard deviation is claculated (excluded are the values with ATF2DELTA109 and EGFR as substrate).
KmP value and equilibrium constant
The Km value for the product is assumed to be similar to but slightly smaller than the the Km value of the substrate because of the similarity of the both species. Therefore the Km value of the substrate is multiplied by 0.95 to gain the one of the product and the uncertainty is increased by increasing the error on Kmsubstrate by 50%.
For information about the equilibrium constant please see here.
References
- ↑ 1.0 1.1 1.2 Haystead T.A. et al. (1992) "Ordered phosphorylation of p42mapk by MAP kinase kinase." (1992) FEBS Lett 306.1, pp.17-22 (pmid:1628739) Cite error: Invalid
<ref>tag; name "Haystead1992" defined multiple times with different content Cite error: Invalid<ref>tag; name "Haystead1992" defined multiple times with different content - ↑ 2.0 2.1 Tian-Rui Xu et al. (2010) "Inferring signaling pathway topologies from multiple perturbation measurements of specific biochemical species." Sci Signal. 3(134):ra20. (pmid:20234003)