Difference between revisions of "Degradation Pathways"
| Line 10: | Line 10: | ||
* [[Decay of PGF2a/exPGF2a |Decay of PGF2a/exPGF2a ]] | * [[Decay of PGF2a/exPGF2a |Decay of PGF2a/exPGF2a ]] | ||
* [[Decay of TXB2/exTXB2 |Decay of TXB2/exTXB2]] | * [[Decay of TXB2/exTXB2 |Decay of TXB2/exTXB2]] | ||
| − | * [[Decay of K6PGF2a to exK6PGF2a |Decay of | + | * [[Decay of K6PGF2a to exK6PGF2a |Decay of 6-keto-PGF1a/ex6-keto-PGF1a ]] |
| − | * [[Decay of PGE2 to exPGE2 |Decay of PGE2 | + | * [[Decay of PGE2 to exPGE2 |Decay of PGE2/exPGE2]] |
| − | * [[Decay of D15PGJ2 to exD15PGJ2 |Decay of | + | * [[Decay of D15PGJ2 to exD15PGJ2 |Decay of 15-deoxy-PGJ2/ex15-deoxy-PGJ2 ]] |
| − | * [[Decay of 5-Oxo-ETE to ex5-Oxo-ETE |Decay of 5- | + | * [[Decay of 5-Oxo-ETE to ex5-Oxo-ETE |Decay of 5-oxo-ETE/ex5-oxo-ETE]] |
| − | * [[Decay of 15-HETE to ex15-HETE |Decay of 15-HETE | + | * [[Decay of 15-HETE to ex15-HETE |Decay of 15-HETE/ex15-HETE]] |
| − | * [[Decay of LTB4 to exLTB4 |Decay of LTB4 | + | * [[Decay of LTB4 to exLTB4 |Decay of LTB4/exLTB4]] |
| − | * [[Decay of LTC4 to exLTC4 |Decay of LTC4 | + | * [[Decay of LTC4 to exLTC4 |Decay of LTC4/exLTC4]] |
| − | * [[Decay of 12-HETE to ex12-HETE |Decay of 12-HETE | + | * [[Decay of 12-HETE to ex12-HETE |Decay of 12-HETE/ex12-HETE]] |
| − | * [[Decay of TXA2 to exTXA2 |Decay of TXA2 | + | * [[Decay of TXA2 to exTXA2 |Decay of TXA2/exTXA2]] |
| − | * [[Decay of PGI2 to exPGI2 |Decay of PGI2 | + | * [[Decay of PGI2 to exPGI2 |Decay of PGI2/exPGI2]] |
| − | * [[Decay of PGH2 to exPGH2 |Decay of PGH2 | + | * [[Decay of PGH2 to exPGH2 |Decay of PGH2/exPGH2 ]] |
| − | * [[Decay of PGD2 to exPGD2 |Decay of PGD2 | + | * [[Decay of PGD2 to exPGD2 |Decay of PGD2/exPGD2]] |
| − | * [[Decay of PGJ2 to exPGJ2 |Decay of PGJ2 | + | * [[Decay of PGJ2 to exPGJ2 |Decay of PGJ2/exPGJ2]] |
| − | * [[Decay of 12-HPETE to ex12-HPETE |Decay of 12-HPETE | + | * [[Decay of 12-HPETE to ex12-HPETE |Decay of 12-HPETE/ex12-HPETE ]] |
| − | * [[Decay of 15-HPETE to ex15-HPETE |Decay of 15-HPETE | + | * [[Decay of 15-HPETE to ex15-HPETE |Decay of 15-HPETE/ex15-HPETE ]] |
| − | * [[Decay of 5-HPETE to ex5-HPETE |Decay of 5-HPETE | + | * [[Decay of 5-HPETE to ex5-HPETE |Decay of 5-HPETE/ex5-HPETE ]] |
| − | * [[Decay of 5-HETE to ex5-HETE |Decay of 5-HETE | + | * [[Decay of 5-HETE to ex5-HETE |Decay of 5-HETE/ex5-HETE ]] |
| − | * [[Decay of LTA4 to exLTA4 |Decay of LTA4 | + | * [[Decay of LTA4 to exLTA4 |Decay of LTA4/exLTA4 ]] |
| − | * [[Decay of AA to exAA|Decay of AA | + | * [[Decay of AA to exAA|Decay of AA/exAA ]] |
| − | * [[Decay of 15-Keto-PGE2 to ex15-Keto-PGE2 |Decay of 15-Keto-PGE2 | + | * [[Decay of 15-Keto-PGE2 to ex15-Keto-PGE2 |Decay of 15-Keto-PGE2/ex15-Keto-PGE2 ]] |
| − | * [[Decay of 3,4-Dihydro-15-Keto-PGE2 to ex3,4-Dihydro-15-Keto-PGE2 |Decay of 3,4-Dihydro-15-Keto-PGE2 | + | * [[Decay of 3,4-Dihydro-15-Keto-PGE2 to ex3,4-Dihydro-15-Keto-PGE2 |Decay of 3,4-Dihydro-15-Keto-PGE2/ex3,4-Dihydro-15-Keto-PGE2 ]] |
|} | |} | ||
Revision as of 14:50, 16 May 2019
Upon being transported out of the cell, the eicosanoids accumulate in the interstitial fluid, which for simplicity is referred to as the extracellular compartment in the model. A decay constant was included for each extracellular metabolite to represent degradation. To describe the breaking down of metabolites an irreversible mass action rate law was used for reactions 43-64. The half life of each eicosanoid was initially assumed as 24 hours, but will be made metabolite specific when all of the values have been collected.
Pseudo-first order reactions.
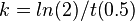
| Reaction # | Species | Half Life (min) | Rate constant(min -1) | Notes | Reference |
|---|---|---|---|---|---|
| 44 | exPGF2a | 900 ± 492 | 0.001 ± 0.001 | Study performed in decidual stromal cells and macrophages in culture. | [1] |
| 45 | exTXB2 | 20 to 30 | 0.035 to 0.023 | Quoted in a textbook(https://books.google.co.uk/books?id=_9kEeTjyJdMC&pg=PA864&lpg=PA864&dq=half+life+txa2&source=bl&ots=2OTF4Mh2Jk&sig=hu79GprliUcW4QE_Zm79islesOA&hl=en&sa=X&ved=0ahUKEwj0oo2sgfjOAhXLIcAKHcaPDHQQ6AEIRjAI#v=onepage&q=half%20life%20txa2&f=false) with no ref. | |
| 46 | exTXA2 | 0.333 | 2.079 | Quoted in a textbook(https://books.google.co.uk/books?id=_9kEeTjyJdMC&pg=PA864&lpg=PA864&dq=half+life+txa2&source=bl&ots=2OTF4Mh2Jk&sig=hu79GprliUcW4QE_Zm79islesOA&hl=en&sa=X&ved=0ahUKEwj0oo2sgfjOAhXLIcAKHcaPDHQQ6AEIRjAI#v=onepage&q=half%20life%20txa2&f=false) with no ref. | |
| 47 | ex6-KETO-PGF1A | 30 | 0.0231 | Human Plasma | [2] |
| 48 | exPGI2 | 3 | 0.231 | [3] | |
| 49 | exPGE2 | 528 ± 204 | 0.001 ± 0.003 | Study performed in decidual stromal cells and macrophages in culture. | [1] |
| 50 | ex15-DEOXY-PGJ2 | 720 | 0.001 | Dehydration of PGD2 to ultimatley 15d-PGJ2 occurs with a half life of about 12 hours in the presense of albumin (protien found in blood). | [4] |
| 51 | exPGJ2 | ||||
| 52 | exPGD2 | 1.5 - 1.6 | 0.462 to 0.433 | Human brain | [5] |
| 30 | 0.023 | Human plasma | [6] | ||
| 53 | exPGH2 | 5 | 0.139 | Quoted on supplier page (http://www.enzolifesciences.com/BML-PH002/prostaglandin-h2/) | |
| 54 | ex5-OXO-ETE | 11 | 0.064 | Study half life of 15-OXO-ETE in in R15L Cells | [7] |
| 55 | ex5-HETE | ||||
| 56 | exLTB4 | 0.47 ± 0.02 to 0.63 ± 0.04 | 1.475 ± 34.657 to 1.100 ± 17.329 | Rabbit, Immunoreactive LTB4 | [8] |
| 57 | exLTC4 | ||||
| 58 | exLTA4 | 0.05 | 13.863 | 37 degrees C | [9] |
| 59 | ex5-HPETE | ||||
| 60 | ex15-HETE | 21 | 0.0331 | Study in R15L Cells | [7] |
| 61 | ex15-HPETE | ||||
| 62 | ex12-HETE | 180 | 0.004 | "During the first 2 min., the half-life of 12-HETE was 0.9 s, which implies
a fast clearance of the compound from the circulation. However, during the subsequent half-hour the estimated half-life was 3 min. and increased dramatically at the interval of time from 30 to 60 min. (t1/2 around 3 h)." |
[10] |
| 63 | ex12-HPETE | 0.5 | 1.386 | [11] | |
| 64 | exAA | 240 to 660 | 0.003 to 0.001 | [12] |
References
- ↑ 1.0 1.1 O. Ishihara, "Differences of metabolism of prostaglandin E2 and F2 alpha by decidual stromal cells and macrophages in culture." Eicosanoids. 1991;4(4):203-7.
- ↑ Ylikorkala, "Measurement of 6-keto-prostaglandin E1 alpha in human plasma with radioimmunoassay: effect of prostacyclin infusion." Prostaglandins Med. 1981 Apr;6(4):427-36.
- ↑ Cawello W., "Metabolism and pharmacokinetics of prostaglandin E1 administered by intravenous infusion in human subjects." Eur J Clin Pharmacol. 1994;46(3):275-7.
- ↑ F. Fitzpatrick, "Albumin-catalyzed metabolism of prostaglandin D2. Identification of products formed in vitro." J Biol Chem. 1983 Oct 10;258(19):11713-8.
- ↑ Suzuki F. "Transport of prostaglandin D2 into brain." Brain Res. 1986 Oct 22;385(2):321-8.
- ↑ R. Schuligoi. "PGD2 metabolism in plasma: Kinetics and relationship with bioactivity on DP1 and CRTH2 receptors" Biochemical Pharmacology, Volume 74, Issue 1, 30 June 2007, Pages 107-117
- ↑ 7.0 7.1 Cong W., "15-oxo-Eicosatetraenoic Acid, a Metabolite of Macrophage 15-Hydroxyprostaglandin Dehydrogenase That Inhibits Endothelial Cell Proliferation" Mol Pharmacol. 2009 Sep; 76(3): 516–525.
- ↑ Marleau S., "Metabolic disposition of leukotriene B4 (LTB4) and oxidation-resistant analogues of LTB4 in conscious rabbits." Br J Pharmacol. 1994 Jun;112(2):654-8.
- ↑ Zimmer J., "Fatty acid binding proteins stabilize leukotriene A4 competition with arachidonic acid but not other lipoxygenase products" November 2004 The Journal of Lipid Research, 45, 2138-2144.
- ↑ Dadaian M., "12-hydroxyeicosatetraenoic acid is a long-lived substance in the rabbit circulation." Prostaglandins Other Lipid Mediat. 1998 Jan;55(1):3-25.
- ↑ J. Maclouf, "Stimulation of leukotriene biosynthesis in human blood leukocytes by platelet-derived 12-hydroperoxy-icosatetraenoic acid" (1982) Proc. Natl. Acad. Sci. U. S. A. 79, 6042-6046
- ↑ Vinge E., "Arachidonic acid-induced platelet aggregation and prostanoid formation in whole blood in relation to plasma concentration of indomethacin." Eur J Clin Pharmacol. 1985;28(2):163-9.