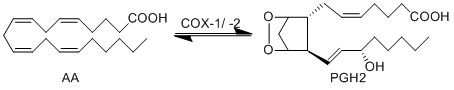
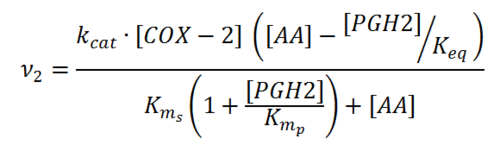Transformation of AA to PGH2
The COX enzyme possesses two isoforms, COX-1 and COX-2, of which both produce bioactive eicosanoids. The two isoforms have nearly identical active site residues (Simmons et al. 2004), but COX-1 is a constitutive enzyme, whereas COX-2 is an inducible enzyme. Both enzymes add molecular oxygen to arachidonic acid, generating an endoperoxide species, PGH2, by catalysing the two-step reaction of cycloooxygenation and oxygenation, followed by a hydroperoxide reduction.
PGH2 possesses a short life time due to the formation of a reactive functional group. The endoperoxide undergoes rapid isomerisation reactions, catalysed by a series of synthase enzymes (PGES, PGDS, PGIS, PGFS and TXAS). The resultant species of the isomerisation reactions are PGs (PGE2, PGD2, PGJ2, deoxy-PGJ2 and PGF2α), TXs (TXA2 and TXB2) and prostacyclin (PGI2), all of which have varying effects on the immune system.
Contents
Reaction
Chemical equation
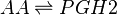
Rate equation
Enzyme Parameters
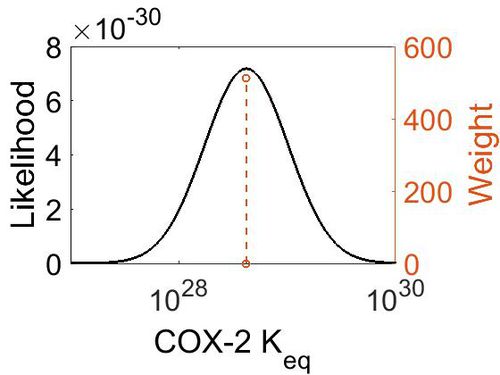
Keq
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| (-30) | kcal/mol | Unspecified | Calculations with a Gaussian98 suite of programs
Enzyme: COX (Unspecific) Substrate: Arachidonate Temperature: 298.15 K Pressure: 1 bar |
[1] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 4.18E+28 | 1.00E+01 | 6.67E+01 | 8.90E-01 |
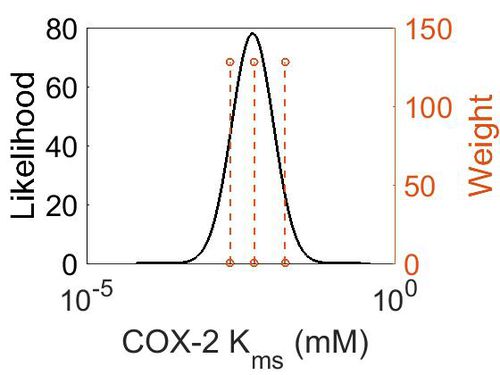
Kms
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 1.62E-02 ± 0.22E-02 | 
|
Human | Expression Vector: Embryonic kidney cells
Enzyme: Cyclooxygenase-2 pH: Not specified Temperature: Not specified |
[2] |
| 5.14E-03 ± 2.90E-04 | mM | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[3] |
| 2.1E-03 ± 4.00E-04 | mM | Human | Expression Vector: E. Coli
Enzyme: Wild Type Cyclooxygenase-2 Enzyme pH: 8.5 Temperature:30 °C |
[4] |
| 2.1E-03 ± 4.00E-04 | mM | Human | Expression Vector: Baculuvirus
Enzyme: Wild Type Cycloxygenase-2 pH: 7.2 Temperature: 30 °C 100 uM arachidonate substrate, |
[5] |
| Mode (mM) | Confidence Interval | Location parameter (μ) | Scale parameter (σ) |
|---|---|---|---|
| 4.90E-03 | 5.70E+00 | -4.72E+00 | 7.77E-01 |
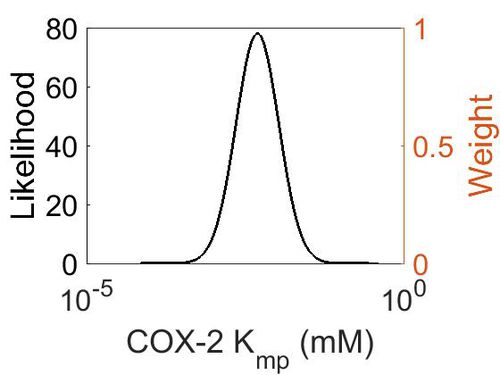
Kmp (Dependent parameter)
This is a “Dependent parameter”, meaning that the log-normal distribution for this parameter was calculated using multivariate distributions (discussed in Protocol for defining informative priors for ensemble modelling in systems biology). As a result, no confidence interval factor or literature values were cited for this parameter.
| Mode | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 4.70E-03 | -4.75E+00 | 7.87E-01 |
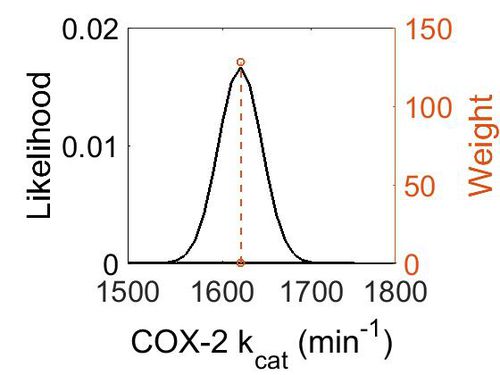
kcat
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 1620 ± 24 | per minute | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[3] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.62E+03 | 1.01E+00 | 7.39E+00 | 1.48E-02 |
Enzyme concentration
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 13.7 | 
|
Human | Expression Vector: Platlet
Enzyme: Cyclooxygenase-2 (PGTS2) pH: 7.5 Temperature: 37 °C |
[6] |
| 4.11 | 
|
Human | Expression Vector: Stomach
Enzyme: Cyclooxygenase-2 (PGTS2) pH: 7.5 Temperature: 37 °C |
[7] |
| 1.49 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: Cyclooxygenase-2 (PGTS2) pH: 7.5 Temperature: 37 °C |
[7] |
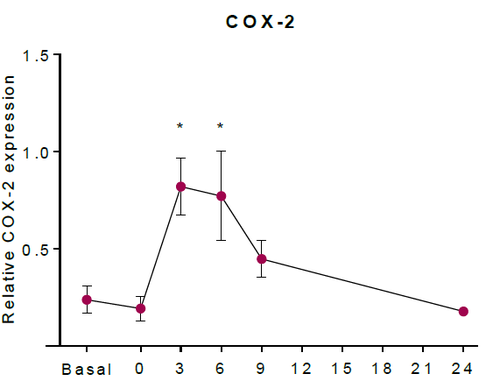
The abundance of COX-2 (ppm) was converted to COX-2 (mM). As a result, the concentration of COX-2 in unstimulated tissue was estimated as 2.27 x10-5 mM. The upregulation of COX-2 in HaCaT keratinocytes was estimated using western blotting in (Kiezel-Tsugunova, 2017), figure * shows an example. Using this information, in silico experiments which included an upregulation of COX-2, included the concentration of COX-2 reaching 100 times higher concentration than the estimated unstimulated concentration. Therefore, the COX-2 induction event includes the concentration of COX-2 eventually reaching a concentration of 2.27 x10-3 mM after 6h post irradiation.
COX Enzyme Parameters with other substrates
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 9.45E-03 ± 7.30E-04 | mM | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 522 ± 12.6 | per minute | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 3.68E-02 ± 4.25E-03 | mM | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 207 ± 9 | per minute | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[3] |
References
- ↑ P. Silva, "A theoretical study of radical-only and combined radical/carbocationic mechanisms of arachidonic acid cyclooxygenation by prostaglandin H synthase" Theor Chem Acc (2003) 110: 345
- ↑ S.F. Kim Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science 2005,310(5756):1966-70)
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 [www.ncbi.nlm.nih.gov/pubmed/20463020 Vecchio A. J. "Structural basis of fatty acid substrate binding to cyclooxygenase-2." J. Biol. Chem. 285 22152-63 (2010)]
- ↑ [http://pubs.acs.org/doi/pdf/10.1021/bi035717o Rogge C. "Identification of Tyr504 as an Alternative Tyrosyl Radical Site in Human Prostaglandin H Synthase-2" Biochemistry 2004, 43, 1560-1568]
- ↑ [http://www.jbc.org/content/279/6/4084.full.pdf Bambai B. "Role of Asn-382 and Thr-383 in Activation and Inactivation of Human Prostaglandin H Synthase Cyclooxygenase Catalysis" February 6, 2004 The Journal of Biological Chemistry, 279, 4084-4092.]
- ↑ M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ 7.0 7.1 [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587]