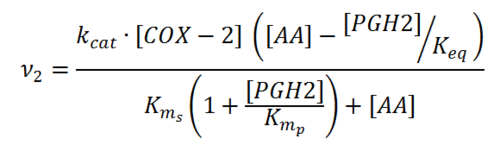Difference between revisions of "Transformation of AA to PGH2"
(→COX-2 Enzyme Parameters) |
|||
| (19 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
[[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | [[Welcome to the In-Silico Model of Cutaneous Lipids Wiki | Return to overview]] | ||
| − | The COX | + | Cyclooxygenase, COX, also known as prostaglandin endoperoxide H (PGH) synthase, has a broad substrate specificity and is reported to metabolise AA and other fatty acids into prostanoids such as PGs, TXs and prostacyclin <ref>Tsai, Al Wei, C. Baek, H. K. Kulmacz, R. J. Van Wart, H. E., ''Comparison of peroxidase reaction mechanisms of prostaglandin H synthase-1 containing heme and mangano protoporphyrin IX'', J Biol Chem (1997), 272, 8885-94.</ref><ref>Kulmacz, R. J. van der Donk, W. A. Tsai, A. L. , ''Comparison of the properties of prostaglandin H synthase-1 and -2'', Prog Lipid Res (2003), 42, 377-404</ref>. The preferred substrate is AA for both isoforms of COX, COX-1 and COX-2. The COX-1 isoform is constitutively expressed, whereas COX-2 expression is typically negligible in normal cells <ref>Gurram, B. Zhang, S. Li, M. Li, H. Xie, Y. Cui, H. Du, J. Fan, J. Wang, J. Peng, X. , ''Celecoxib conjugated fluorescent probe for identification and discrimination of cyclooxygenase-2 enzyme in cancer cells'', Anal Chem (2018), 90, 5187-5193.</ref> but can be induced in response to inflammatory stimuli, hormones, calcium and growth factors<ref>Fletcher, B. S. Kujubu, D. A. Perrin, D. M. Herschman, H. R. , ''Structure of the mitogen-inducible TIS10 gene and demonstration that the TIS10-encoded protein is a functional prostaglandin G/H synthase'', J Biol Chem (1992), 267, 4338-44.</ref><ref>Xie, W. L. Chipman, J. G. Robertson, D. L. Erikson, R. L. Simmons, D. L. , ''Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing'', Proc Natl Acad Sci U S A (1991), 88, 2692-6.</ref><ref>Ristimaki, A. Garfinkel, S. Wessendorf, J. Maciag, T. Hla, T. , ''Induction of cyclooxygenase-2 by interleukin-1 alpha. Evidence for post-transcriptional regulation'', J Biol Chem (1994), 269, 11769-75.</ref><ref>name, ''title'', journal (year)</ref><ref>Fu, J. Y. Masferrer, J. L. Seibert, K. Raz, A. Needleman, P., ''The induction and suppression of prostaglandin H2 synthase (cyclooxygenase) in human monocytes'', J Biol Chem (1990), 265, 16737-40.</ref><ref>Kujubu, D. A. Reddy, S. T. Fletcher, B. S. Herschman, H. R., ''Expression of the protein product of the prostaglandin synthase-2/TIS10 gene in mitogen-stimulated Swiss 3T3 cells'', J Biol Chem (1993), 268, 5425-30.</ref> <ref>Wang, D. An, S. J. Wang, W. H. McGiff, J. C. Ferreri, N. R., ''CaR-mediated COX-2 expression in primary cultured mTAL cells'', Am J Physiol Renal Physiol (2001), 281, F658-64.)</ref>. Interestingly, basal expression of COX-2 has been found to occur in the kidney, central nervous system, female reproductive organs and stomach <ref>Obermoser, V. Baecker, D. Schuster, C. Braun, V. Kircher, B. Gust, R. , ''Chlorinated cobalt alkyne complexes derived from acetylsalicylic acid as new specific antitumor agents'', Dalton Trans, 47, 4341-4351.</ref><ref>Kirkby, N. S. Chan, M. V. Zaiss, A. K. Garcia-Vaz, E. Jiao, J. Berglund, L. M. Verdu, E. F. Ahmetaj-Shala, B. Wallace, J. L. Herschman, H. R. Gomez, M. F. Mitchell, J. A., ''Systematic study of constitutive cyclooxygenase-2 expression: Role of NF-kappaB and NFAT transcriptional pathways'', Proc Natl Acad Sci U S A (2016), 113, 434-9.</ref>, and frequent expression of COX-2 can be found in the tumorigenic nests of most cancers <ref>Gurram, B. Zhang, S. Li, M. Li, H. Xie, Y. Cui, H. Du, J. Fan, J. Wang, J. Peng, X. , ''Celecoxib conjugated fluorescent probe for identification and discrimination of cyclooxygenase-2 enzyme in cancer cells'', Anal Chem (2018), 90, 5187-5193.</ref><ref>Raj, V. Bhadauria, A. S. Singh, A. K. Kumar, U. Rai, A. Keshari, A. K. Kumar, P. Kumar, D. Maity, B. Nath, S. Prakash, A. Ansari, K. M. Jat, J. L. Saha, S. , ''Novel 1,3,4-thiadiazoles inhibit colorectal cancer via blockade of IL-6/COX-2 mediated JAK2/STAT3 signals as evidenced through data-based mathematical modeling'', Cell, 135, 216-26.</ref>. |
| + | |||
| + | Both isoforms of COX catalyse a two-step reaction of cycloooxygenation and oxygenation, followed by a hydroperoxide reduction. The cyclooxygenase reaction occurs in the hydrophobic channel within the core of the protein and generates an unstable peroxide intermediate, PGG2. The subsequent peroxidase reaction produces PGH2 and occurs at the heme-containing active site near the protein surface. The two step reaction results in the insertion of molecular oxygen across the C-9 and C-11 double bonds. Both isoforms of COX are heme-containing glycoproteins, that are membrane-bound to the endoplasmic reticulum (ER) and function as homodimers <ref>Smith, W. L. DeWitt, D. L. Garavito, R. M., ''Cyclooxygenases: structural, cellular, and molecular biology'', Annu Rev Biochem (2000), 69, 145-82.</ref><ref>van der Donk, W. A. Tsai, A. L. Kulmacz, R. J., ''The cyclooxygenase reaction mechanism'', Biochemistry, 41, 15451-8.</ref><ref>Rouzer, C. A. Marnett, L. J., ''Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases'', Chem Rev, 103, 2239-304.</ref><ref>Xiao, G. Chen, W. Kulmacz, R. J., ''Comparison of structural stabilities of prostaglandin H synthase-1 and -2'', J Biol Chem (1998), 273, 6801-11.</ref>. Interestingly, in certain tissues COX-1 has been found to colocalises preferentially, but not exclusively, with TXAS, PGFS and cPGES, whereas COX-2 has been found to colocalise with PGIS and mPGES in certain tissues <ref>Miyata, A. Yokoyama, C. Ihara, H. Bandoh, S. Takeda, O. Takahashi, E. Tanabe, T., ''Characterization of the human gene (TBXAS1) encoding thromboxane synthase'', Eur J Biochem (1994), 224, 273-9.</ref><ref>Ullrich, V. Zou, M. H. Bachschmid, M. , ''New physiological and pathophysiological aspects on the thromboxane A(2)-prostacyclin regulatory system'', Biochim Biophys Acta (2001), 1532, 1-14.</ref><ref>Smyth, E. M. Grosser, T. Wang, M. Yu, Y. FitzGerald, G. A., ''Prostanoids in health and disease'', J Lipid Res, 50 Suppl, S423-8.</ref><ref>Smith, W. L. DeWitt, D. L. Garavito, R. M., ''Cyclooxygenases: structural, cellular, and molecular biology'', Annu Rev Biochem (2000), 69, 145-82.</ref><ref>Ueno, N. Murakami, M. Tanioka, T. Fujimori, K. Tanabe, T. | ||
| + | Urade, Y. Kudo, I. , ''Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2'', J Biol Chem (2001), 276, 34918-27.</ref>. | ||
| − | |||
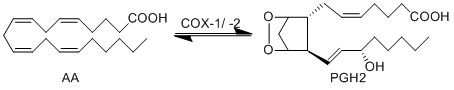
== Reaction == | == Reaction == | ||
| Line 16: | Line 18: | ||
[[File:R02.PNG|center|500px]] | [[File:R02.PNG|center|500px]] | ||
| − | == | + | == Enzyme Parameters == |
| − | + | === K<sub>eq</sub> === | |
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
|+ style="text-align: left;" | Gibbs Free Energy Change | |+ style="text-align: left;" | Gibbs Free Energy Change | ||
| Line 25: | Line 27: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 35: | Line 38: | ||
Temperature: 298.15 K | Temperature: 298.15 K | ||
Pressure: 1 bar | Pressure: 1 bar | ||
| + | | 64 | ||
|<ref name="Silva2003”>[http://link.springer.com/article/10.1007/s00214-003-0476-9 P. Silva, "A theoretical study of radical-only and combined radical/carbocationic mechanisms of arachidonic acid cyclooxygenation by prostaglandin H synthase" Theor Chem Acc (2003) 110: 345]</ref> | |<ref name="Silva2003”>[http://link.springer.com/article/10.1007/s00214-003-0476-9 P. Silva, "A theoretical study of radical-only and combined radical/carbocationic mechanisms of arachidonic acid cyclooxygenation by prostaglandin H synthase" Theor Chem Acc (2003) 110: 345]</ref> | ||
|- | |- | ||
| Line 40: | Line 44: | ||
|} | |} | ||
| − | + | {| class="wikitable" | |
| − | {|class="wikitable | + | |+ style="text-align: left;" | Description of the COX-2 Keq distribution |
| − | |+ style="text-align: left;" | COX- | + | ! Mode !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) |
|- | |- | ||
| − | + | | 4.18E+28 || 1.00E+01 || 6.67E+01 || 8.90E-01 | |
| − | |||
| − | |||
| − | |||
| − | |||
|- | |- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
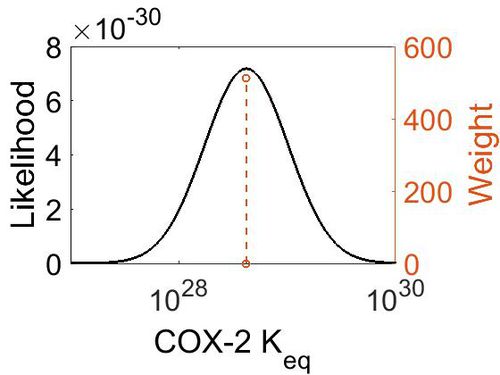
| − | + | [[Image:Cox-2_keq.jpg|none|thumb|500px|The estimated probability distribution for COX-2 Keq. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | | | ||
| − | | | ||
| − | | | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== K<sub>ms</sub> === | === K<sub>ms</sub> === | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | Literature | + | |+ style="text-align: left;" | Literature Information |
|- | |- | ||
! Value | ! Value | ||
| Line 170: | Line 63: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 179: | Line 73: | ||
pH: Not specified | pH: Not specified | ||
Temperature: Not specified | Temperature: Not specified | ||
| + | |128 | ||
|<ref name="Kim2005"> [http://www.ncbi.nlm.nih.gov/pubmed/16373578 S.F. Kim Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2.'' Science 2005,310(5756):1966-70)]</ref> | |<ref name="Kim2005"> [http://www.ncbi.nlm.nih.gov/pubmed/16373578 S.F. Kim Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2.'' Science 2005,310(5756):1966-70)]</ref> | ||
|- | |- | ||
| Line 189: | Line 84: | ||
Temperature: 37 °C | Temperature: 37 °C | ||
1–200 µM of substrate. | 1–200 µM of substrate. | ||
| + | |128 | ||
|<ref name="Vecchoi2010"> [www.ncbi.nlm.nih.gov/pubmed/20463020 Vecchio A. J. "Structural basis of fatty acid substrate binding to cyclooxygenase-2." J. Biol. Chem. 285 22152-63 (2010)]</ref> | |<ref name="Vecchoi2010"> [www.ncbi.nlm.nih.gov/pubmed/20463020 Vecchio A. J. "Structural basis of fatty acid substrate binding to cyclooxygenase-2." J. Biol. Chem. 285 22152-63 (2010)]</ref> | ||
|- | |- | ||
| Line 198: | Line 94: | ||
pH: 8.5 | pH: 8.5 | ||
Temperature:30 °C | Temperature:30 °C | ||
| + | |64 | ||
|<ref name="Rogge2003"> [http://pubs.acs.org/doi/pdf/10.1021/bi035717o Rogge C. "Identification of Tyr504 as an Alternative Tyrosyl Radical Site in Human | |<ref name="Rogge2003"> [http://pubs.acs.org/doi/pdf/10.1021/bi035717o Rogge C. "Identification of Tyr504 as an Alternative Tyrosyl Radical Site in Human | ||
Prostaglandin H Synthase-2" Biochemistry 2004, 43, 1560-1568]</ref> | Prostaglandin H Synthase-2" Biochemistry 2004, 43, 1560-1568]</ref> | ||
| Line 208: | Line 105: | ||
pH: 7.2 | pH: 7.2 | ||
Temperature: 30 °C | Temperature: 30 °C | ||
| − | 100 uM arachidonate substrate, | + | 100 uM arachidonate substrate, |
| + | |128 | ||
|<ref name="Bambai2004"> [http://www.jbc.org/content/279/6/4084.full.pdf Bambai B. "Role of Asn-382 and Thr-383 in Activation and Inactivation of | |<ref name="Bambai2004"> [http://www.jbc.org/content/279/6/4084.full.pdf Bambai B. "Role of Asn-382 and Thr-383 in Activation and Inactivation of | ||
Human Prostaglandin H Synthase Cyclooxygenase Catalysis" February 6, 2004 The Journal of Biological Chemistry, 279, 4084-4092.]</ref> | Human Prostaglandin H Synthase Cyclooxygenase Catalysis" February 6, 2004 The Journal of Biological Chemistry, 279, 4084-4092.]</ref> | ||
| Line 216: | Line 114: | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Description of the COX-2 Kms distribution |
|- | |- | ||
! Mode (mM) !! Confidence Interval !! Location parameter (μ) !! Scale parameter (σ) | ! Mode (mM) !! Confidence Interval !! Location parameter (μ) !! Scale parameter (σ) | ||
| Line 223: | Line 121: | ||
|} | |} | ||
| − | [[ | + | |
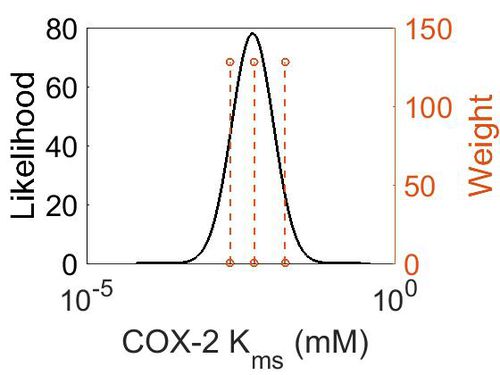
| + | [[Image:5.jpg|none|thumb|500px|The estimated probability distribution for COX-2 Kms. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
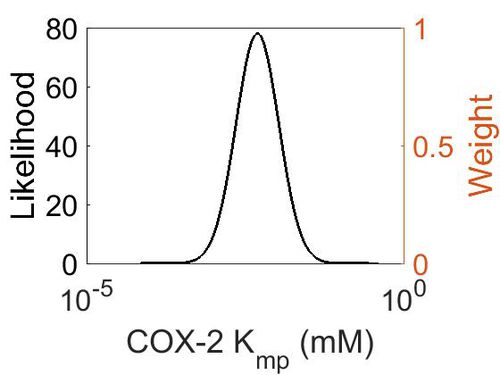
=== K<sub>mp</sub> (Dependent parameter) === | === K<sub>mp</sub> (Dependent parameter) === | ||
| + | This is a “Dependent parameter”, meaning that the log-normal distribution for this parameter was calculated using multivariate distributions (discussed in [[Quantification of parameter uncertainty | Protocol for defining informative priors for ensemble modelling in systems biology]]). As a result, no confidence interval factor or literature values were cited for this parameter. | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Description of the COX-2 Kmp distribution |
! Mode !! Location parameter (µ) !! Scale parameter (σ) | ! Mode !! Location parameter (µ) !! Scale parameter (σ) | ||
|- | |- | ||
| Line 235: | Line 135: | ||
|} | |} | ||
| − | [[ | + | [[Image:6.jpg|none|thumb|500px|The estimated probability distribution for COX-2 Kmp. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] |
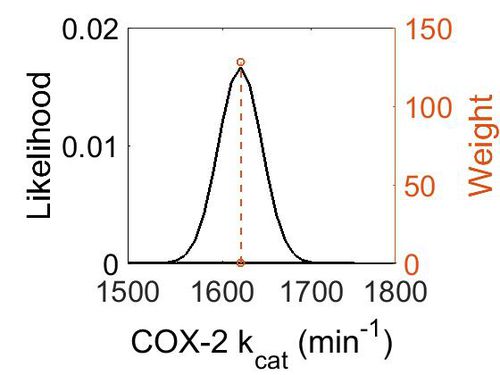
=== k<sub>cat</sub> === | === k<sub>cat</sub> === | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | + | |+ style="text-align: left;" | Literature Information | |
|- | |- | ||
! Value | ! Value | ||
| Line 246: | Line 146: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 256: | Line 157: | ||
Temperature: 37 °C | Temperature: 37 °C | ||
1–200 µM of substrate. | 1–200 µM of substrate. | ||
| + | |128 | ||
|<ref name="Vecchoi2010"> [www.ncbi.nlm.nih.gov/pubmed/20463020 Vecchio A. J. "Structural basis of fatty acid substrate binding to cyclooxygenase-2." J. Biol. Chem. 285 22152-63 (2010)]</ref> | |<ref name="Vecchoi2010"> [www.ncbi.nlm.nih.gov/pubmed/20463020 Vecchio A. J. "Structural basis of fatty acid substrate binding to cyclooxygenase-2." J. Biol. Chem. 285 22152-63 (2010)]</ref> | ||
|- | |- | ||
| Line 261: | Line 163: | ||
{| class="wikitable" | {| class="wikitable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | Description of the COX-2 k<sub>cat</sub> distribution |
! Mode (min-1) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ! Mode (min-1) !! Confidence Interval !! Location parameter (µ) !! Scale parameter (σ) | ||
|- | |- | ||
| Line 267: | Line 169: | ||
|} | |} | ||
| − | [[ | + | [[Image:Cox2_kcat.jpg|none|thumb|500px|The estimated probability distribution for COX-2 kcat. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] |
| − | |||
=== Enzyme concentration === | === Enzyme concentration === | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | + | |+ style="text-align: left;" | Literature Information | |
| − | |||
|- | |- | ||
! Value | ! Value | ||
| Line 280: | Line 180: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 289: | Line 190: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
| Line 298: | Line 200: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | ||
human proteome'' Nature, 2014 509, 582–587]</ref> | human proteome'' Nature, 2014 509, 582–587]</ref> | ||
| Line 308: | Line 211: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | ||
human proteome'' Nature, 2014 509, 582–587]</ref> | human proteome'' Nature, 2014 509, 582–587]</ref> | ||
| Line 315: | Line 219: | ||
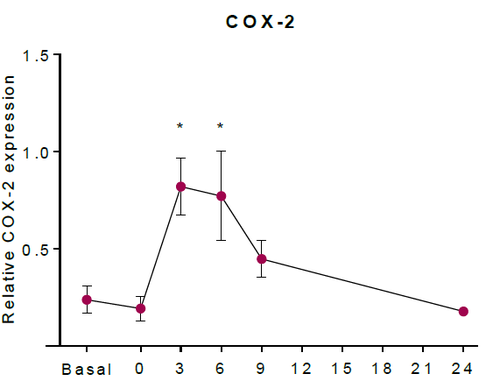
The abundance of COX-2 (ppm) was converted to COX-2 (mM). As a result, the concentration of COX-2 in unstimulated tissue was estimated as 2.27 x10-5 mM. The upregulation of COX-2 in HaCaT keratinocytes was estimated using western blotting in (Kiezel-Tsugunova, 2017), figure * shows an example. Using this information, in silico experiments which included an upregulation of COX-2, included the concentration of COX-2 reaching 100 times higher concentration than the estimated unstimulated concentration. Therefore, the COX-2 induction event includes the concentration of COX-2 eventually reaching a concentration of 2.27 x10-3 mM after 6h post irradiation. | The abundance of COX-2 (ppm) was converted to COX-2 (mM). As a result, the concentration of COX-2 in unstimulated tissue was estimated as 2.27 x10-5 mM. The upregulation of COX-2 in HaCaT keratinocytes was estimated using western blotting in (Kiezel-Tsugunova, 2017), figure * shows an example. Using this information, in silico experiments which included an upregulation of COX-2, included the concentration of COX-2 reaching 100 times higher concentration than the estimated unstimulated concentration. Therefore, the COX-2 induction event includes the concentration of COX-2 eventually reaching a concentration of 2.27 x10-3 mM after 6h post irradiation. | ||
| − | [[ | + | [[Image:MKT_COX-2.png|none|thumb|500px| The relative expression of COX-2 protein in HaCaT keratinocytes (Kiezel-Tsugunova, 2017).]] |
== COX Enzyme Parameters with other substrates == | == COX Enzyme Parameters with other substrates == | ||
Latest revision as of 15:41, 20 August 2019
Cyclooxygenase, COX, also known as prostaglandin endoperoxide H (PGH) synthase, has a broad substrate specificity and is reported to metabolise AA and other fatty acids into prostanoids such as PGs, TXs and prostacyclin [1][2]. The preferred substrate is AA for both isoforms of COX, COX-1 and COX-2. The COX-1 isoform is constitutively expressed, whereas COX-2 expression is typically negligible in normal cells [3] but can be induced in response to inflammatory stimuli, hormones, calcium and growth factors[4][5][6][7][8][9] [10]. Interestingly, basal expression of COX-2 has been found to occur in the kidney, central nervous system, female reproductive organs and stomach [11][12], and frequent expression of COX-2 can be found in the tumorigenic nests of most cancers [13][14].
Both isoforms of COX catalyse a two-step reaction of cycloooxygenation and oxygenation, followed by a hydroperoxide reduction. The cyclooxygenase reaction occurs in the hydrophobic channel within the core of the protein and generates an unstable peroxide intermediate, PGG2. The subsequent peroxidase reaction produces PGH2 and occurs at the heme-containing active site near the protein surface. The two step reaction results in the insertion of molecular oxygen across the C-9 and C-11 double bonds. Both isoforms of COX are heme-containing glycoproteins, that are membrane-bound to the endoplasmic reticulum (ER) and function as homodimers [15][16][17][18]. Interestingly, in certain tissues COX-1 has been found to colocalises preferentially, but not exclusively, with TXAS, PGFS and cPGES, whereas COX-2 has been found to colocalise with PGIS and mPGES in certain tissues [19][20][21][22][23].
Contents
Reaction
Chemical equation
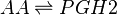
Rate equation
Enzyme Parameters
Keq
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| (-30) | kcal/mol | Unspecified | Calculations with a Gaussian98 suite of programs
Enzyme: COX (Unspecific) Substrate: Arachidonate Temperature: 298.15 K Pressure: 1 bar |
64 | [24] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 4.18E+28 | 1.00E+01 | 6.67E+01 | 8.90E-01 |
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 1.62E-02 ± 0.22E-02 | 
|
Human | Expression Vector: Embryonic kidney cells
Enzyme: Cyclooxygenase-2 pH: Not specified Temperature: Not specified |
128 | [25] |
| 5.14E-03 ± 2.90E-04 | mM | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
128 | [26] |
| 2.1E-03 ± 4.00E-04 | mM | Human | Expression Vector: E. Coli
Enzyme: Wild Type Cyclooxygenase-2 Enzyme pH: 8.5 Temperature:30 °C |
64 | [27] |
| 2.1E-03 ± 4.00E-04 | mM | Human | Expression Vector: Baculuvirus
Enzyme: Wild Type Cycloxygenase-2 pH: 7.2 Temperature: 30 °C 100 uM arachidonate substrate, |
128 | [28] |
| Mode (mM) | Confidence Interval | Location parameter (μ) | Scale parameter (σ) |
|---|---|---|---|
| 4.90E-03 | 5.70E+00 | -4.72E+00 | 7.77E-01 |
Kmp (Dependent parameter)
This is a “Dependent parameter”, meaning that the log-normal distribution for this parameter was calculated using multivariate distributions (discussed in Protocol for defining informative priors for ensemble modelling in systems biology). As a result, no confidence interval factor or literature values were cited for this parameter.
| Mode | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 4.70E-03 | -4.75E+00 | 7.87E-01 |
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 1620 ± 24 | per minute | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
128 | [26] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.62E+03 | 1.01E+00 | 7.39E+00 | 1.48E-02 |
Enzyme concentration
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 13.7 | 
|
Human | Expression Vector: Platlet
Enzyme: Cyclooxygenase-2 (PGTS2) pH: 7.5 Temperature: 37 °C |
1024 | [29] |
| 4.11 | 
|
Human | Expression Vector: Stomach
Enzyme: Cyclooxygenase-2 (PGTS2) pH: 7.5 Temperature: 37 °C |
1024 | [30] |
| 1.49 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: Cyclooxygenase-2 (PGTS2) pH: 7.5 Temperature: 37 °C |
1024 | [30] |
The abundance of COX-2 (ppm) was converted to COX-2 (mM). As a result, the concentration of COX-2 in unstimulated tissue was estimated as 2.27 x10-5 mM. The upregulation of COX-2 in HaCaT keratinocytes was estimated using western blotting in (Kiezel-Tsugunova, 2017), figure * shows an example. Using this information, in silico experiments which included an upregulation of COX-2, included the concentration of COX-2 reaching 100 times higher concentration than the estimated unstimulated concentration. Therefore, the COX-2 induction event includes the concentration of COX-2 eventually reaching a concentration of 2.27 x10-3 mM after 6h post irradiation.
COX Enzyme Parameters with other substrates
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 9.45E-03 ± 7.30E-04 | mM | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[26] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 522 ± 12.6 | per minute | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[26] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 3.68E-02 ± 4.25E-03 | mM | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[26] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 207 ± 9 | per minute | Mouse | Expression Vector: Baculovirus (Insect Cell)
Enzyme: COX-2 pH: 8.0 Temperature: 37 °C 1–200 µM of substrate. |
[26] |
References
- ↑ Tsai, Al Wei, C. Baek, H. K. Kulmacz, R. J. Van Wart, H. E., Comparison of peroxidase reaction mechanisms of prostaglandin H synthase-1 containing heme and mangano protoporphyrin IX, J Biol Chem (1997), 272, 8885-94.
- ↑ Kulmacz, R. J. van der Donk, W. A. Tsai, A. L. , Comparison of the properties of prostaglandin H synthase-1 and -2, Prog Lipid Res (2003), 42, 377-404
- ↑ Gurram, B. Zhang, S. Li, M. Li, H. Xie, Y. Cui, H. Du, J. Fan, J. Wang, J. Peng, X. , Celecoxib conjugated fluorescent probe for identification and discrimination of cyclooxygenase-2 enzyme in cancer cells, Anal Chem (2018), 90, 5187-5193.
- ↑ Fletcher, B. S. Kujubu, D. A. Perrin, D. M. Herschman, H. R. , Structure of the mitogen-inducible TIS10 gene and demonstration that the TIS10-encoded protein is a functional prostaglandin G/H synthase, J Biol Chem (1992), 267, 4338-44.
- ↑ Xie, W. L. Chipman, J. G. Robertson, D. L. Erikson, R. L. Simmons, D. L. , Expression of a mitogen-responsive gene encoding prostaglandin synthase is regulated by mRNA splicing, Proc Natl Acad Sci U S A (1991), 88, 2692-6.
- ↑ Ristimaki, A. Garfinkel, S. Wessendorf, J. Maciag, T. Hla, T. , Induction of cyclooxygenase-2 by interleukin-1 alpha. Evidence for post-transcriptional regulation, J Biol Chem (1994), 269, 11769-75.
- ↑ name, title, journal (year)
- ↑ Fu, J. Y. Masferrer, J. L. Seibert, K. Raz, A. Needleman, P., The induction and suppression of prostaglandin H2 synthase (cyclooxygenase) in human monocytes, J Biol Chem (1990), 265, 16737-40.
- ↑ Kujubu, D. A. Reddy, S. T. Fletcher, B. S. Herschman, H. R., Expression of the protein product of the prostaglandin synthase-2/TIS10 gene in mitogen-stimulated Swiss 3T3 cells, J Biol Chem (1993), 268, 5425-30.
- ↑ Wang, D. An, S. J. Wang, W. H. McGiff, J. C. Ferreri, N. R., CaR-mediated COX-2 expression in primary cultured mTAL cells, Am J Physiol Renal Physiol (2001), 281, F658-64.)
- ↑ Obermoser, V. Baecker, D. Schuster, C. Braun, V. Kircher, B. Gust, R. , Chlorinated cobalt alkyne complexes derived from acetylsalicylic acid as new specific antitumor agents, Dalton Trans, 47, 4341-4351.
- ↑ Kirkby, N. S. Chan, M. V. Zaiss, A. K. Garcia-Vaz, E. Jiao, J. Berglund, L. M. Verdu, E. F. Ahmetaj-Shala, B. Wallace, J. L. Herschman, H. R. Gomez, M. F. Mitchell, J. A., Systematic study of constitutive cyclooxygenase-2 expression: Role of NF-kappaB and NFAT transcriptional pathways, Proc Natl Acad Sci U S A (2016), 113, 434-9.
- ↑ Gurram, B. Zhang, S. Li, M. Li, H. Xie, Y. Cui, H. Du, J. Fan, J. Wang, J. Peng, X. , Celecoxib conjugated fluorescent probe for identification and discrimination of cyclooxygenase-2 enzyme in cancer cells, Anal Chem (2018), 90, 5187-5193.
- ↑ Raj, V. Bhadauria, A. S. Singh, A. K. Kumar, U. Rai, A. Keshari, A. K. Kumar, P. Kumar, D. Maity, B. Nath, S. Prakash, A. Ansari, K. M. Jat, J. L. Saha, S. , Novel 1,3,4-thiadiazoles inhibit colorectal cancer via blockade of IL-6/COX-2 mediated JAK2/STAT3 signals as evidenced through data-based mathematical modeling, Cell, 135, 216-26.
- ↑ Smith, W. L. DeWitt, D. L. Garavito, R. M., Cyclooxygenases: structural, cellular, and molecular biology, Annu Rev Biochem (2000), 69, 145-82.
- ↑ van der Donk, W. A. Tsai, A. L. Kulmacz, R. J., The cyclooxygenase reaction mechanism, Biochemistry, 41, 15451-8.
- ↑ Rouzer, C. A. Marnett, L. J., Mechanism of free radical oxygenation of polyunsaturated fatty acids by cyclooxygenases, Chem Rev, 103, 2239-304.
- ↑ Xiao, G. Chen, W. Kulmacz, R. J., Comparison of structural stabilities of prostaglandin H synthase-1 and -2, J Biol Chem (1998), 273, 6801-11.
- ↑ Miyata, A. Yokoyama, C. Ihara, H. Bandoh, S. Takeda, O. Takahashi, E. Tanabe, T., Characterization of the human gene (TBXAS1) encoding thromboxane synthase, Eur J Biochem (1994), 224, 273-9.
- ↑ Ullrich, V. Zou, M. H. Bachschmid, M. , New physiological and pathophysiological aspects on the thromboxane A(2)-prostacyclin regulatory system, Biochim Biophys Acta (2001), 1532, 1-14.
- ↑ Smyth, E. M. Grosser, T. Wang, M. Yu, Y. FitzGerald, G. A., Prostanoids in health and disease, J Lipid Res, 50 Suppl, S423-8.
- ↑ Smith, W. L. DeWitt, D. L. Garavito, R. M., Cyclooxygenases: structural, cellular, and molecular biology, Annu Rev Biochem (2000), 69, 145-82.
- ↑ Ueno, N. Murakami, M. Tanioka, T. Fujimori, K. Tanabe, T. Urade, Y. Kudo, I. , Coupling between cyclooxygenase, terminal prostanoid synthase, and phospholipase A2, J Biol Chem (2001), 276, 34918-27.
- ↑ P. Silva, "A theoretical study of radical-only and combined radical/carbocationic mechanisms of arachidonic acid cyclooxygenation by prostaglandin H synthase" Theor Chem Acc (2003) 110: 345
- ↑ S.F. Kim Inducible nitric oxide synthase binds, S-nitrosylates, and activates cyclooxygenase-2. Science 2005,310(5756):1966-70)
- ↑ 26.0 26.1 26.2 26.3 26.4 26.5 [www.ncbi.nlm.nih.gov/pubmed/20463020 Vecchio A. J. "Structural basis of fatty acid substrate binding to cyclooxygenase-2." J. Biol. Chem. 285 22152-63 (2010)]
- ↑ [http://pubs.acs.org/doi/pdf/10.1021/bi035717o Rogge C. "Identification of Tyr504 as an Alternative Tyrosyl Radical Site in Human Prostaglandin H Synthase-2" Biochemistry 2004, 43, 1560-1568]
- ↑ [http://www.jbc.org/content/279/6/4084.full.pdf Bambai B. "Role of Asn-382 and Thr-383 in Activation and Inactivation of Human Prostaglandin H Synthase Cyclooxygenase Catalysis" February 6, 2004 The Journal of Biological Chemistry, 279, 4084-4092.]
- ↑ M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ 30.0 30.1 [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587]