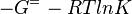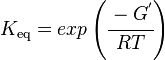Difference between revisions of "Kinetic Model of Monoterpenoid Biosynthesis Wiki"
Aliah.hawari (talk | contribs) (→List of kinetic parameter values) |
Aliah.hawari (talk | contribs) (→Description of the model) |
||
| Line 12: | Line 12: | ||
rect 537 959 639 1001 [[Limonene-3-hydroxylase (L3H)]] | rect 537 959 639 1001 [[Limonene-3-hydroxylase (L3H)]] | ||
rect 180 962 279 998 [[Limonene-6-Hydroxylase (L6H)]] | rect 180 962 279 998 [[Limonene-6-Hydroxylase (L6H)]] | ||
| + | |||
desc bottom-right | desc bottom-right | ||
</imagemap> | </imagemap> | ||
Revision as of 15:12, 16 March 2016
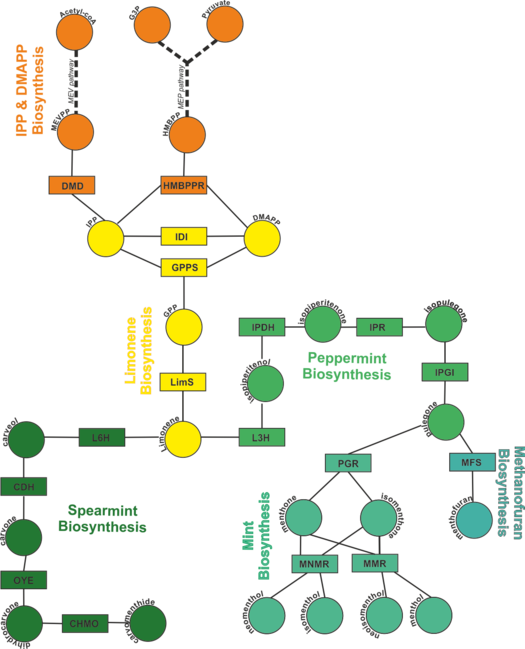
This wiki page describes the construction and simulation of a kinetic model of Monoterpenoid Biosynthesis.
Contents
Monoterpenoid Biosynthesis
Description of the model
To find out how each enzyme in the network is modelled, click on the rectangles in the figure on the left. Alternatively, you can click the enzyme names from the list below.
IPP and DMAPP Biosynthesis
Diphosphomelvalonate decarboxylase (DMD)
Hydroxymethylbutenyl diphosphate reductase (HMBPPR)
Limonene Biosynthesis
Geranyl diphosphate synthase (GPPS)
Peppermint Biosynthesis
Isopiperitenol Dehydrogenase (IPDH)
Isopiperitenol Reductase (IPR)
Mint Biosynthesis
Spearmint Biosynthesis
Methanofuran Biosynthesis
Reversible Michaelis-Menten equation
All reactions in this model are described using reversible Michaelis-Menten equation.
Kinetic Parameters
Strategies for Estimating Kinetic Parameter Values
Using Equilibrium Constant (Keq) in the reversible Michaelis-Menten equation
Using the equilibrium constant in the reversible Michaelis-Menten reaction reduces the need to obtain or estimate Vmaxreverse parameter value, which is often not available in literature.
Using the Haldane relationship, the equilibrium constant (Keq) can be written as:
- Failed to parse (Cannot store math image on filesystem.): {\color{Red}K_\mathrm{eq}} = \frac{Vmax_\mathrm{forward} * Km_\mathrm{product} }{Vmax_\mathrm{reverse} * Km_\mathrm{substrate}}
The reversible Michaelis-Menten rate equation can then be rewritten to contain the equilibrium constant (Keq) instead of using the ratio of the Vmax values.
- Failed to parse (Cannot store math image on filesystem.): v_\mathrm{reaction} = Vmax_\mathrm{forward} * \cfrac {\cfrac{[S]}{Km_\mathrm{substrate}} * \left ( 1 - \cfrac {[P]}{[S]* \color{red}K_\mathrm{eq}} \right )} {1 + \cfrac {[S]}{Km_\mathrm{substrate}} + \cfrac {[P]}{Km_\mathrm{product}}}
where [S] and [P] corresponds to the concentration of the substrate and product respectively.
Calculating the Equilibrium Constant (Keq)
The equilibrium constant can be calculated from the ratio of the forward and reverse reaction rates as described in the above section, given that the values for these rates are known. Unfortunately, the information for these values are often difficult to find, especially for reverse reaction rates (Vmaxreverse).
As an alternative, the equlibrium constant, Keq, can also be calculated from the Gibbs free energy of a reaction, ΔGr, using the Van't Hoff isotherm equation:
and by dividing both sides of the equation with RT, and later take the exponents of both sides, the Keq can be calculated by this equation:
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change |
| R | Gas constant with a value of 8.31 JK-1mol-1 |
| T | Temperature which is always expressed in kelvin |
List of kinetic parameter values
Lists of values retrieved from published and unpublished sources for the kinetics of all the enzymes included in this study can be found here.
Data gathering progress:
| Enzyme | Pathway | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 01_GPPS | Limonene Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 01_L3H | Peppermint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] | ||
| 01_L6H | Spearmint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] | ||
| 01_MFS | Methanofuran Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 01_PGR | Mint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 02_CDH | Spearmint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 02_IPDH | Peppermint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 02_LimSynth | Limonene Biosynthesis | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] | |||
| 02_MNMR (menthone) | Mint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 02a_IPDH | Spearmint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 03_IPR | Peppermint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 03_MMR (menthone) | Mint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 03_OYE | Spearmint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 04_CHMO | Spearmint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 04_IPGI | Peppermint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 04_MNMR (isomenthone) | Mint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| enzyme | Pathway | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
| 04_MNMR (isomenthone) | Mint Biosynthesis | Keq | VmaxF | KmSub1 | KmSub2 | [Sub1] | [Sub2] | VmaxR | KmProd1 | KmProd2 | [Prod1] | [Prod2] |
Abbreviations
List of abbreviations used in this study can be found here
Back to the main model page.