Difference between revisions of "ATP-Binding Cassette Transporters"
(→Parameters) |
(→Reaction) |
||
| Line 14: | Line 14: | ||
== Reaction == | == Reaction == | ||
| − | + | {|width ="80%" | |
| + | | | ||
| + | * [[Transformation of PGF2a to exPGF2a |Transformation of PGF2a to exPGF2a ]] | ||
| + | * [[Transformation of TXB2 to exTXB2 |Transformation of TXB2 to exTXB2]] | ||
| + | * [[Transformation of K6PGF2a to exK6PGF2a |Transformation of K6PGF2a to exK6PGF2a ]] | ||
| + | * [[Transformation of PGE2 to exPGE2 |Transformation of PGE2 to exPGE2]] | ||
| + | * [[Transformation of D15PGJ2 to exD15PGJ2 |Transformation of D15PGJ2 to exD15PGJ2 ]] | ||
| + | * [[Transformation of 5-Oxo-ETE to ex5-Oxo-ETE |Transformation of 5-Oxo-ETE to ex5-Oxo-ETE]] | ||
| + | * [[Transformation of 15-HETE to ex15-HETE |Transformation of 15-HETE to ex15-HETE]] | ||
| + | * [[Transformation of LTB4 to exLTB4 |Transformation of LTB4 to exLTB4]] | ||
| + | * [[Transformation of LTC4 to exLTC4 |Transformation of LTC4 to exLTC4]] | ||
| + | * [[Transformation of 12-HETE to ex12-HETE |Transformation of 12-HETE to ex12-HETE]] | ||
| + | * [[Transformation of TXA2 to exTXA2 |Transformation of TXA2 to exTXA2]] | ||
| + | * [[Transformation of PGI2 to exPGI2 |Transformation of PGI2 to exPGI2]] | ||
| + | * [[Transformation of PGH2 to exPGH2 |Transformation of PGH2 to exPGH2 ]] | ||
| + | * [[Transformation of PGD2 to exPGD2 |Transformation of PGD2 to exPGD2]] | ||
| + | * [[Transformation of PGJ2 to exPGJ2 |Transformation of PGJ2 to exPGJ2]] | ||
| + | * [[Transformation of 12-HPETE to ex12-HPETE |Transformation of 12-HPETE to ex12-HPETE ]] | ||
| + | * [[Transformation of 15-HPETE to ex15-HPETE |Transformation of 15-HPETE to ex15-HPETE ]] | ||
| + | * [[Transformation of 5-HPETE to ex5-HPETE |Transformation of 5-HPETE to ex5-HPETE ]] | ||
| + | * [[Transformation of 5-HETE to ex5-HETE |Transformation of 5-HETE to ex5-HETE ]] | ||
| + | * [[Transformation of LTA4 to exLTA4 |Transformation of LTA4 to exLTA4 ]] | ||
| + | * [[Transformation of AA to exAA|Transformation of AA to exAA ]] | ||
| + | * [[Transformation of 15-Keto-PGE2 to ex15-Keto-PGE2 |Transformation of 15-Keto-PGE2 to ex15-Keto-PGE2 ]] | ||
| + | * [[Transformation of 3,4-Dihydro-15-Keto-PGE2 to ex3,4-Dihydro-15-Keto-PGE2 |Transformation of 3,4-Dihydro-15-Keto-PGE2 to ex3,4-Dihydro-15-Keto-PGE2 ]] | ||
| + | |} | ||
==Chemical equation== | ==Chemical equation== | ||
Revision as of 14:46, 26 September 2017
When a cell produces eicosanoids they are immediately transported into the extracellular compartment, as they are cytotoxic \cite{Pompeia2002}. An ATP-binding cassette (ABC) transporter has been assumed as the method of transportation across the cellular membrane for reactions 22-43.
An ABC transporter was assumed as the export transporter of lipids because it is widely reported as transporting organic molecules (ref). Due to the increased amount of free energy provided by the ATP hydrolysis reaction, the transporter is able to relocate molecules against a concentration gradient. The model only contains an inward facing transporter, although in reality there is also an outward facing variant.
In the literature a prostaglandin transporter is reported (Schuster2015), but due to the lack of kinetic analysis of the transporter, the ABC transporter was modelled. We assume that rate of transportation across the membrane may be similar between transporters, however once specific transporter kinetics become available, the model will be updated.
The rate law was designed to encompass the basic principles of an ATP transporter, e.g. the consumption of the internal lipid is directly proportional to amount of transporters. The ratio of substrate and product describes how the gradient would affect the rate of transportation, for example if the concentration of the external lipid equalled that of the internal, transportation would cease. The final term in the equation is the driving force for the transportation, calculated using Gibbs free energy of ATP hydrolysis, the gas constant, temperature and the concentrations of ATP and ADP. As the reformation of ATP is considerably faster than the lipid transporter the ratio was assumed to remain constant.
Contents
Reaction
Chemical equation
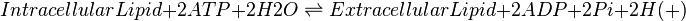
Rate equation
Parameters
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.0109 ± 0.0041 | mM | Human | Substrate: LTC4
pH 7.0, 37°C, recombinant MRP2, using 4mM MgATP and 5g of isolated membranes. |
[1] |
| 0.0000366 ± 0.0000038 | mM | Human | Substrate LTC4, 37 °C, 4 mM ATP, | [2] |
| 5.3E-3 ± 2.6E-3 | mM | Human | 4 mM ATP, 37°C, 5–10 ml of membrane vesicle suspension (30
mg protein). |
[3] |
| 0.1954 ± 0.0612 | mM | Human | pH 7.0, 37°C, recombinant MRP2,
Substrate: 17beta-estradiol 17-(beta-D-glucuronide) |
[1] |
| 0.1097 ± 0.0391 | mM | Human | pH 7.0, 37°C, recombinant MRP2,
Substrate: Estrone glucuronide |
[1] |
| 0.0207 | mM | Human | pH 7.4, 37°C, Substrate: Progesterone, Expression vector: Human HepG2 hepatoma | [4] |
No data available on the turnover of ABCC4 transporter therefore based upon estimates made by http://book.bionumbers.org/what-are-the-rates-of-membrane-transporters/
| Value | Units | Transporter | Notes | Reference |
|---|---|---|---|---|
| 3.5 | min-1 | Glucose transporter ptsI | Organism: Bacteria Escherichia coli
Back of the envelope calculation by BioNumbers: http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=102931 |
[5] |
| 0.33 - 0.83 | min-1 | Rate of transport of H+/Lactose transporter | Organism: Bacteria Escherichia coli
Back of the envelop calculation on bionumbers http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=103159 |
[6] |
| 3.28 | min-1 | Catalytic Rate of transporter HXT7 | Organism: Budding yeast Saccharomyces cerevisiae
Back of the envelope calculation http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=101737 |
[7] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.00019 ± 1.96e-5 | mmol/min/mg protein | Human | Substrate: LTC4
pH 7.0, 37°C, recombinant MRP2, using 4mM MgATP and 5g of isolated membranes. |
[1] |
| 1.25e-7 ± 1.2e-8 | mmol/min/mg | Human | Substrate LTC4, 37 °C, 4 mM ATP, | [2] |
| 2.02e-8 ± 5.9e-9 | mmol/mg/min | Human | 4 mM ATP, 37°C, 5–10 ml of membrane vesicle suspension (30
mg protein). |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 5.15 | ppm | Human | Expression Vector: Stomach
Enzyme: ABCC4 pH: 7.5 Temperature: 37 °C |
[8] |
| 5.94 | ppm | Human | Expression Vector: Lung
Enzyme: ABCC4 pH: 7.5 Temperature: 37 °C |
[9] |
| 2.66 | ppm | Human | Expression Vector: Gut
Enzyme: ABCC4 pH: 7.5 Temperature: 37 °C |
[9] |
References
- ↑ 1.0 1.1 1.2 1.3 Yasunaga M. "Molecular cloning and functional characterization of cynomolgus monkey multidrug resistance-associated protein 2 (MRP2) Eur. J. Pharm. Sci. 35, 326-334 (2008)
- ↑ 2.0 2.1 Zeng H. "Transport of amphipathic anions by human multidrug resistance protein 3 J. Biol. Chem. 275, 34166-34172 (2000) Cite error: Invalid
<ref>tag; name "Mao2000" defined multiple times with different content - ↑ 3.0 3.1 "Functional reconstitution of substrate transport by purified multidrug resistance protein MRP1 (ABCC1) in phospholipid vesicles Cancer Res. 60, 4779-4784 (2000)
- ↑ W. Fong "Schisandrol A from Schisandra chinensis Reverses P-Glycoprotein-Mediated Multidrug Resistance by Affecting Pgp-Substrate Complexes Planta Med 2007; 73(3): 212-220
- ↑ Waygood EB Enzyme I of the phosphoenolpyruvate: sugar phosphotransferase system of Escherichia coli. Purification to homogeneity and some properties.Can J Biochem. 1980 Jan;58(1):40-8.
- ↑ Naftalin R. J. ELactose permease H+-lactose symporter: mechanical switch or Brownian ratchet?Biophys J. 2007 May 15;92(10):3474-91. Epub 2007 Feb 26.
- ↑ Ye L. Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiaeYeast. 2001 Sep 30;18(13):1257-67.
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ 9.0 9.1 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581