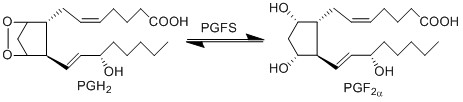
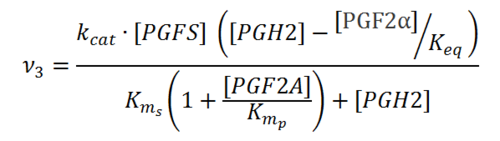Transformation of PGH2 to PGF2α
The isomerisation of PGH2 to PGF2a is performed by PGFS. This reaction results in the formation of two hydroxyl groups at C9 and C11. This reaction takes place in the cytosol of cells and the cofactor is NADPH. PGFS is found to also catalyse the conversion of aldehydes and ketones to PGF2a, for example the conversion of PGD2 to PGF2a.
PGF2α has been found to be produced by keratinocyte cells in vitro (Rhodes, Gledhill et al. 2009). This species is synthesised from PGH2 via the prostaglandin F synthase (PGFS) or the PGE 9-ketoreductase enzymes. Upon production this species have been associated with pigmentation, inflammation and hair growth in the skin (Scott, Leopardi et al. 2004, Scott, Fricke et al. 2007). The species has various inflammatory roles such as a pro-inflammatory effect upon the system by facilitating pain transmission (Murata, Ushikubi et al. 1997) and also an anti-inflammatory effect by inhibiting lymphocyte and endothelial cell interactions (Della Bella, Molteni et al. 2001).
Contents
Reaction
Chemical equation
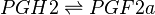
Rate equation
Parameters
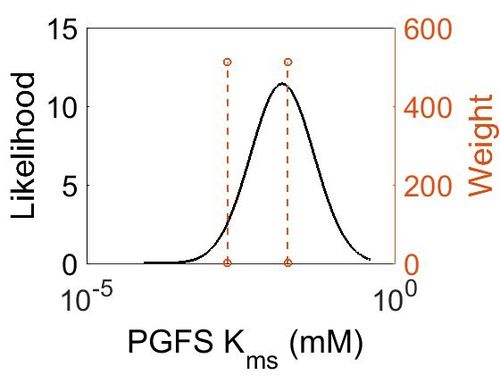
Kms
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 1.90E-03 ± 1.5E-03 | 
|
Human | Placental Aldose Reductase (AKR1B1), E. Coli, pH 7, 37°C | [1] |
| 1.80E-02 | 
|
Human | Lung PGF2a Synthase (AKR1C3), E. Coli, pH 7, 37°C | [1] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.33E-02 | 3.81E+00 | -3.45E+00 | 9.33E-01 |
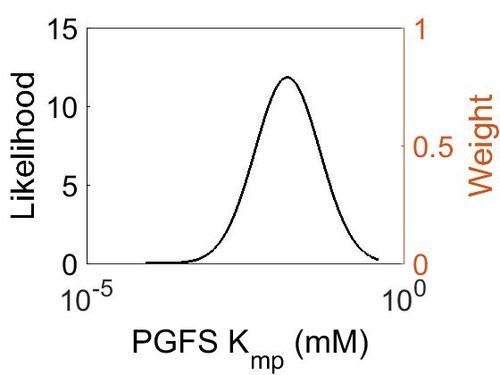
Kmp
In this model some parameters are referred as “Dependent parameters”, meaning that the log-normal distribution for that parameter was calculated using multivariate distributions (discussed in Protocol for defining informative priors for ensemble modelling in systems biology). As a result, no confidence interval factor or literature values were cited for the Dependent parameters.
| Mode | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.46E-02 | -2.86E+00 | 1.17E+00 |
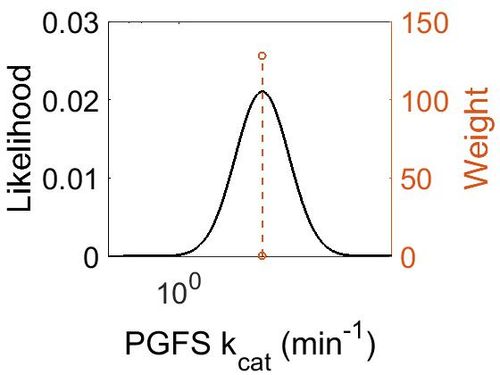
kcat
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 14.9 | per minute | Mouse/Swine | Prostamide/PGF Synthase Purified
from Swine Brain and of Murine Enzyme Expressed in E. coli |
[2] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.48E+01 | 1.10E+00 | 2.71E+00 | 9.49E-02 |
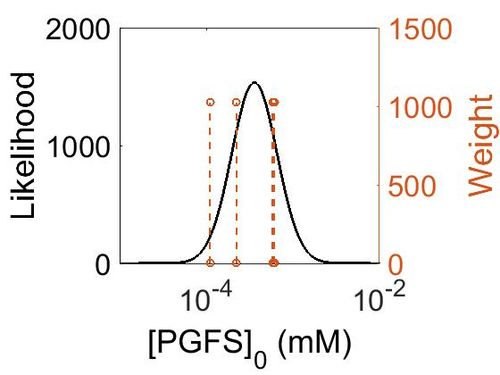
Enzyme concentration
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 105 | 
|
Human | Expression Vector: Platelet
Enzyme: PGFS pH: 7.5 Temperature: 37 °C |
[3] |
| 19.9 | 
|
Human | Expression Vector: Lung
Enzyme: PGFS pH: 7.5 Temperature: 37 °C |
[4] |
| 40.3 | 
|
Human | Expression Vector: Esophagus
Enzyme: PGFS pH: 7.5 Temperature: 37 °C |
[4] |
| 110 | 
|
Human | Expression Vector: Oral Cavity
Enzyme: PGFS pH: 7.5 Temperature: 37 °C |
[4] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 6.41E+01 | 2.05E+00 | 4.53E+00 | 6.06E-01 |
References
- ↑ 1.0 1.1 Z. Kabututu, M. Manin, Prostaglandin F2alpha synthase activities of aldo-keto reductase 1B1, 1B3 and 1B7. J Biochem 2009,145(2):161-8)
- ↑ Moriuchi H., Molecular characterization of a novel type of prostamide/prostaglandin F synthase, belonging to the thioredoxin-like superfamily. J Biol Chem. 2008 Jan 11;283(2):792-801.)
- ↑ M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ 4.0 4.1 4.2 M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587