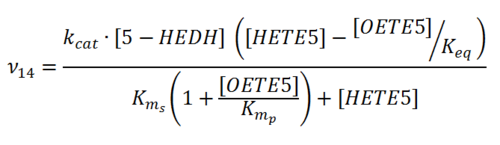Difference between revisions of "Transformation of 5-HETE to 5-OXO-ETE"
(→Parameters) |
|||
| Line 26: | Line 26: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 32: | Line 33: | ||
| Unkown | | Unkown | ||
| Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE. | | Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE. | ||
| + | |64 | ||
| <ref name="Steinhilber"> [https://books.google.co.uk/books?id=mEYWDAAAQBAJ&pg=PA188&lpg=PA188&dq=5-HEDH+Km&source=bl&ots=dwhpDJqBhn&sig=jgK1WqNLlylOaNedgGdwNBQN7TM&hl=en&sa=X&ved=0ahUKEwiw_pCykbfPAhVDD8AKHZu2BnwQ6AEIHDAA#v=onepage&q=5-HEDH%20Km&f=false Lipoxygenases in Inflammation - edited by Dieter Steinhilber]</ref> | | <ref name="Steinhilber"> [https://books.google.co.uk/books?id=mEYWDAAAQBAJ&pg=PA188&lpg=PA188&dq=5-HEDH+Km&source=bl&ots=dwhpDJqBhn&sig=jgK1WqNLlylOaNedgGdwNBQN7TM&hl=en&sa=X&ved=0ahUKEwiw_pCykbfPAhVDD8AKHZu2BnwQ6AEIHDAA#v=onepage&q=5-HEDH%20Km&f=false Lipoxygenases in Inflammation - edited by Dieter Steinhilber]</ref> | ||
|- | |- | ||
| Line 44: | Line 46: | ||
pH: 7.4 | pH: 7.4 | ||
Temperature: 37 ◦C | Temperature: 37 ◦C | ||
| + | |2048 | ||
| <ref name="ERLEMANN2007"> [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828885/pdf/bj4030157.pdf Karl-Rudolf ERLEMANN, Regulation of 5-hydroxyeicosanoid dehydrogenase activity in | | <ref name="ERLEMANN2007"> [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828885/pdf/bj4030157.pdf Karl-Rudolf ERLEMANN, Regulation of 5-hydroxyeicosanoid dehydrogenase activity in | ||
monocytic cells, Biochem. J. (2007) 403, 157–165]</ref> | monocytic cells, Biochem. J. (2007) 403, 157–165]</ref> | ||
| Line 57: | Line 60: | ||
pH: 7.4 | pH: 7.4 | ||
Temperature: 37 ◦C | Temperature: 37 ◦C | ||
| + | |2048 | ||
| <ref name="Patel2009"> [http://jpet.aspetjournals.org/content/jpet/329/1/335.full.pdf Karl-Pranav Patel, Selectivity of 5-Hydroxyeicosanoid Dehydrogenase | | <ref name="Patel2009"> [http://jpet.aspetjournals.org/content/jpet/329/1/335.full.pdf Karl-Pranav Patel, Selectivity of 5-Hydroxyeicosanoid Dehydrogenase | ||
and Its Inhibition by 5-Hydroxy-Long-Chain Fatty Acids, Journal of Pharmacology and Experimental Therapeutics April 2009, 329 (1) 335-341; ]</ref> | and Its Inhibition by 5-Hydroxy-Long-Chain Fatty Acids, Journal of Pharmacology and Experimental Therapeutics April 2009, 329 (1) 335-341; ]</ref> | ||
| Line 128: | Line 132: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 141: | Line 146: | ||
Substrate: NADPH | Substrate: NADPH | ||
| + | |128 | ||
| B | | B | ||
|} | |} | ||
| Line 161: | Line 167: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 170: | Line 177: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |2048 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | ||
human proteome'' Nature, 2014 509, 582–587]</ref> | human proteome'' Nature, 2014 509, 582–587]</ref> | ||
| Line 180: | Line 188: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | ||
human proteome'' Nature, 2014 509, 582–587]</ref> | human proteome'' Nature, 2014 509, 582–587]</ref> | ||
| Line 190: | Line 199: | ||
pH: 7.5 | pH: 7.5 | ||
Temperature: 37 °C | Temperature: 37 °C | ||
| + | |1024 | ||
|<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
| Line 199: | Line 209: | ||
pH: Unknown | pH: Unknown | ||
Temperature: Unknown | Temperature: Unknown | ||
| + | |2048 | ||
|<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
| Line 221: | Line 232: | ||
! Species | ! Species | ||
! Notes | ! Notes | ||
| + | ! Weight | ||
! Reference | ! Reference | ||
|- | |- | ||
| Line 234: | Line 246: | ||
pH: 7.3 | pH: 7.3 | ||
ionic strength: 0.25 | ionic strength: 0.25 | ||
| + | |64 | ||
|<ref name="MetaCyc”>[https://metacyc.org/META/NEW-IMAGE?type=REACTION&object=RXN-16416 Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | |<ref name="MetaCyc”>[https://metacyc.org/META/NEW-IMAGE?type=REACTION&object=RXN-16416 Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471]</ref> | ||
|} | |} | ||
| Line 245: | Line 258: | ||
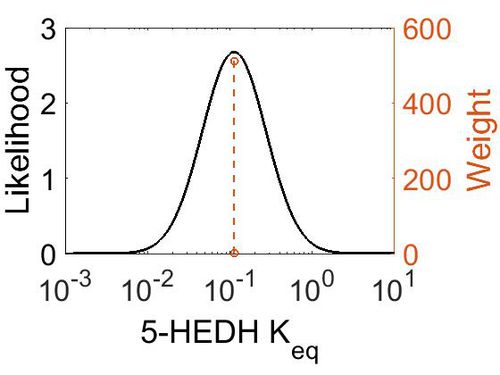
[[Image:48.jpg|none|thumb|500px|The estimated probability distribution for 5-HEDH Keq. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | [[Image:48.jpg|none|thumb|500px|The estimated probability distribution for 5-HEDH Keq. The value and weight of the literature values used to define the distribution are indicated by an orange dashed line. The x axis is plotted on a log-scale. ]] | ||
| − | |||
== Related Reactions == | == Related Reactions == | ||
Revision as of 11:25, 22 May 2019
The metabolism of 5-HETE to 5-Oxo-ETE is catalysed by the enzyme 5-Hydroxyeicosanoid dehydrogenase (5-HEDH). This enzyme acts reversibly and has been reported to be found in eosinophils, monocytes, DC, B-lymphocytes, keratinocytes and platelets (Steinhilber book) *skin (need to check). The product binds with the OXE receptor (Hosoi2002) and is 30-100 more bioactive than 5-HETE. 5-Oxo-ETE is a chemoattractant for neutrophils and eosinophils.
Contents
Reaction
Chemical equation
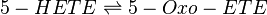
Rate equation
Parameters
Note that there was limited availability of kinetic data for this specific enzyme, therefore parameters for general alcohol dehydrogenase (ADH) was used when non were available.
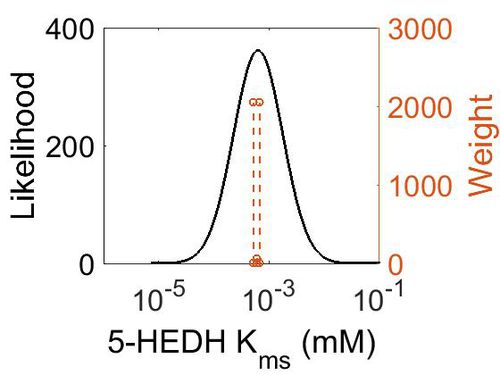
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
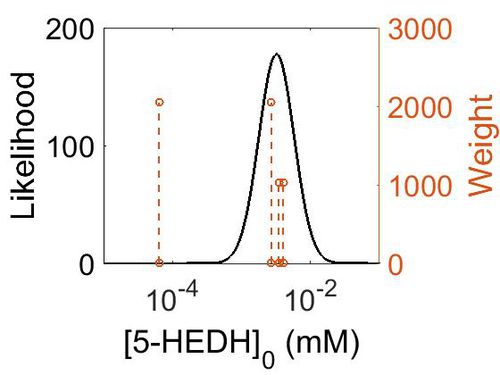
| 0.0006 | mM | Unkown | Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE. | 64 | [1] |
| 0.00067 | mM | Human Cell lines | Follows a ping-pong mechanism by binding with NADP+ (Km 139 nM), transforming it to NADPH (inhibited by NADPH (Ki 224 nM)) and releasing it before binding 5S-HETE.
Method: In vitro Organism: Human Expression vector: U937 and HL-60 cell lines Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
2048 | [2] |
| 0.000516 ± 0.00019 | mM | Human Cell lines |
Method: In vitro Organism: Human Expression vector: U937 Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
2048 | [3] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 6.34E-04 | 1.63E+01 | -6.31E+00 | 1.03E+00 |
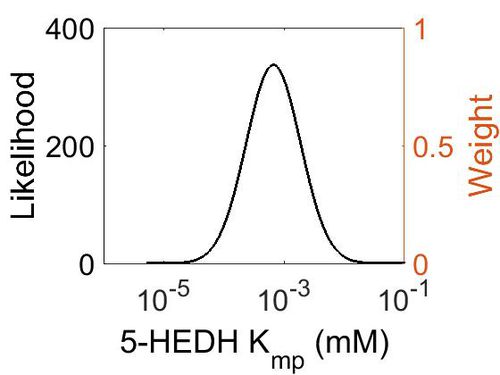
Kmp
This is a “Dependent parameter”, meaning that the log-normal distribution for this parameter was calculated using multivariate distributions (this is discussed in detail here). As a result, no confidence interval factor or literature values were cited for this parameter.
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 1.14E+04 | 1.09E+01 | 1.25E+00 |
Vmax
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.54 ± 0.30 | pmol/ mi mg | Human Cell lines - Neutrophils |
Method: In vitro Organism: Human Expression vector: U937 and HL-60 cell lines Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
[2] |
| 0.40 ± 0.12 | pmol/ mi mg | Human Cell lines - U937 |
Method: In vitro Organism: Human Expression vector: U937 and HL-60 cell lines Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
[2] |
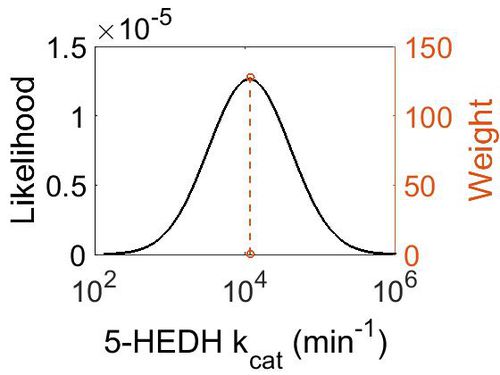
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 11640 ± 840 | min-1 | Yokenella | Method: In vitro
Organism: Human Expression vector: E Coli Enzyme: pH: 6.5 Temperature: 65 ◦C Substrate: NADPH |
128 | B |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.15E+04 | 4.02E+01 | 1.09E+01 | 1.25E+00 |
Enzyme concentration (Using ADH5 abundance data)
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 490 | ppm | Human | Expression Vector: Skin
Enzyme: ADH5 pH: 7.5 Temperature: 37 °C |
2048 | [4] |
| 625 | ppm | Human | Expression Vector: Oral Cavity
Enzyme: ADH5 pH: 7.5 Temperature: 37 °C |
1024 | [4] |
| 728 | ppm | Human | Expression Vector: Esophagus
Enzyme: ADH5 pH: 7.5 Temperature: 37 °C |
1024 | [5] |
| 11.5 | ppm | Human | Expression Vector: Skin
Enzyme: ADH5 pH: Unknown Temperature: Unknown |
2048 | [5] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 4.88E+02 | 6.37E+00 | 7.49E+00 | 1.14E+00 |
Keq
| Gibbs Free energy | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 1.29 | kcal/mol | Arabidopsis thaliana col | Same reaction, no prediction available for 5-HEDH on Metacyc
Estimated Enzyme: ω-hydroxy fatty acid ω-alcohol dehydrogenase Substrate: 18-hydroxyoleate Product: 18-oxo-oleate pH: 7.3 ionic strength: 0.25 |
64 | [6] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.12E-01 | 1.00E+01 | -1.39E+00 | 8.91E-01 |
Related Reactions
References
- ↑ Lipoxygenases in Inflammation - edited by Dieter Steinhilber
- ↑ 2.0 2.1 2.2 [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828885/pdf/bj4030157.pdf Karl-Rudolf ERLEMANN, Regulation of 5-hydroxyeicosanoid dehydrogenase activity in monocytic cells, Biochem. J. (2007) 403, 157–165]
- ↑ [http://jpet.aspetjournals.org/content/jpet/329/1/335.full.pdf Karl-Pranav Patel, Selectivity of 5-Hydroxyeicosanoid Dehydrogenase and Its Inhibition by 5-Hydroxy-Long-Chain Fatty Acids, Journal of Pharmacology and Experimental Therapeutics April 2009, 329 (1) 335-341; ]
- ↑ 4.0 4.1 [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587]
- ↑ 5.0 5.1 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471