Difference between revisions of "Transformation of 5-HETE to 5-OXO-ETE"
(→Parameters) |
|||
| Line 105: | Line 105: | ||
! Reference | ! Reference | ||
|- | |- | ||
| − | | | + | | 11640 ± 840 |
| − | | | + | | min-1 |
| − | | | + | | Yokenella |
| − | | | + | | Method: In vitro |
| + | Organism: Human | ||
| + | Expression vector: E Coli | ||
| + | Enzyme: | ||
| + | pH: 6.5 | ||
| + | Temperature: 65 ◦C | ||
| + | |||
| + | Substrate: NADPH | ||
| B | | B | ||
|} | |} | ||
| + | |||
{|class="wikitable" | {|class="wikitable" | ||
|+ style="text-align: left;" | 5-HEDH Abundance | |+ style="text-align: left;" | 5-HEDH Abundance | ||
Revision as of 13:06, 13 March 2017
The metabolism of 5-HETE to 5-Oxo-ETE is catalysed by the enzyme 5-Hydroxyeicosanoid dehydrogenase (5-HEDH). This enzyme acts reversibly and has been reported to be found in eosinophils, monocytes, DC, B-lymphocytes, keratinocytes and platelets (Steinhilber book) *skin (need to check). The product binds with the OXE receptor (Hosoi2002) and is 30-100 more bioactive than 5-HETE. 5-Oxo-ETE is a chemoattractant for neutrophils and eosinophils.
Contents
Reaction
Chemical equation
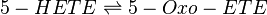
Rate equation
Parameters
Note: Limited availability of kinetic data for specific enzyme, using general alcohol dehydrogenase (ADH) data when non is available.
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.0006 | mM | Unkown | Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE. | [1] |
| 0.00067 | mM | Human Cell lines | Follows a ping-pong mechanism by binding with NADP+ (Km 139 nM), transforming it to NADPH (inhibited by NADPH (Ki 224 nM)) and releasing it before binding 5S-HETE.
Method: In vitro Organism: Human Expression vector: U937 and HL-60 cell lines Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
[2] |
| 0.000516 ± 0.00019 | mM | Human Cell lines |
Method: In vitro Organism: Human Expression vector: U937 Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.54 ± 0.30 | pmol/ mi mg | Human Cell lines - Neutrophils |
Method: In vitro Organism: Human Expression vector: U937 and HL-60 cell lines Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
[2] |
| 0.40 ± 0.12 | pmol/ mi mg | Human Cell lines - U937 |
Method: In vitro Organism: Human Expression vector: U937 and HL-60 cell lines Enzyme:5-HEDH pH: 7.4 Temperature: 37 ◦C |
[2] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 11640 ± 840 | min-1 | Yokenella | Method: In vitro
Organism: Human Expression vector: E Coli Enzyme: pH: 6.5 Temperature: 65 ◦C Substrate: NADPH |
B |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| X | Y | Z | A | B |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 1.29 | kcal/mol | Arabidopsis thaliana col | Same reaction, no prediction available for 5-HEDH on Metacyc
Estimated Enzyme: ω-hydroxy fatty acid ω-alcohol dehydrogenase Substrate: 18-hydroxyoleate Product: 18-oxo-oleate pH: 7.3 ionic strength: 0.25 |
[4] |
Related Reactions
References
- ↑ Lipoxygenases in Inflammation - edited by Dieter Steinhilber
- ↑ 2.0 2.1 2.2 [https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1828885/pdf/bj4030157.pdf Karl-Rudolf ERLEMANN, Regulation of 5-hydroxyeicosanoid dehydrogenase activity in monocytic cells, Biochem. J. (2007) 403, 157–165]
- ↑ [http://jpet.aspetjournals.org/content/jpet/329/1/335.full.pdf Karl-Pranav Patel, Selectivity of 5-Hydroxyeicosanoid Dehydrogenase and Its Inhibition by 5-Hydroxy-Long-Chain Fatty Acids, Journal of Pharmacology and Experimental Therapeutics April 2009, 329 (1) 335-341; ]
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471
