Difference between revisions of "Transformation of 5-HETE to 5-OXO-ETE"
(→Parameters) |
(→Parameters) |
||
| Line 24: | Line 24: | ||
! Reference | ! Reference | ||
|- | |- | ||
| − | | | + | | 0.0006 |
| − | | | + | | mM |
| − | | | + | | Unkown |
| − | | | + | | Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE. |
| − | | | + | | <ref name="Steinhilber"> [https://books.google.co.uk/books?id=mEYWDAAAQBAJ&pg=PA188&lpg=PA188&dq=5-HEDH+Km&source=bl&ots=dwhpDJqBhn&sig=jgK1WqNLlylOaNedgGdwNBQN7TM&hl=en&sa=X&ved=0ahUKEwiw_pCykbfPAhVDD8AKHZu2BnwQ6AEIHDAA#v=onepage&q=5-HEDH%20Km&f=false Lipoxygenases in Inflammation - edited by Dieter Steinhilber]</ref> |
|} | |} | ||
Revision as of 14:05, 30 September 2016
The metabolism of 5-HETE to 5-Oxo-ETE is catalysed by the enzyme 5-Hydroxyeicosanoid dehydrogenase (5-HEDH). This enzyme acts reversibly and has been reported to be found in skin (need to check). The product binds with the OXE receptor (Hosoi2002) and is 30-100 more bioactive than 5-HETE. 5-Oxo-ETE is a chemoattractant for neutrophils and eosinophils.
Reaction
Chemical equation
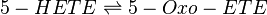
Rate equation
Parameters
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.0006 | mM | Unkown | Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE. | [1] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| X | Y | Z | A | B |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| X | Y | Z | A | B |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| X | Y | Z | A | B |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| X | Y | Z | A | B |
