Difference between revisions of "Formation of homo-dimer R2"
(→Parameters) |
(→Parameters) |
||
| Line 36: | Line 36: | ||
|- | |- | ||
|<math>k^{-}_{6}</math> | |<math>k^{-}_{6}</math> | ||
| − | |<math> | + | |<math>2.12-6.84</math> <ref name="Janin1997"> [http://onlinelibrary.wiley.com/doi/10.1002/(SICI)1097-0134(199706)28:2%3C153::AID-PROT4%3E3.0.CO;2-G/epdf Janin, Joel. ''The kinetics of protein-protein recognition.'' Proteins-Structure Function and Bioinformatics (1997): 153-161.]</ref> <ref name="Northrup1992"> [http://www.pnas.org/content/89/8/3338.full.pdf Northrup S.H. and Erickson H.P. ''Kinetics of protein-protein association explained by Brownian dynamics computer simulation.''PNAS 1992;89(8),3338-3342]</ref> |
|<math>min^{-1}</math> | |<math>min^{-1}</math> | ||
| N/A | | N/A | ||
| − | |According to Northrup et al. the <math>K_{a}</math> of protein-protein bond formations occur in the order of <math>10^{6} M^{-1}s^{-1}</math>. Therefore, this value is used to calculate the dissociation rate for the ScbR homo-dimer formation. Therefore the range of the <math>k^{-}_{3}</math> values is calculated as per <math>k^{-}_{6}=K_{d6} \cdot K_{a}</math>. | + | |According to Northrup et al. the <math>K_{a}</math> of protein-protein bond formations occur in the order of <math>10^{6} M^{-1}s^{-1}</math>. Therefore, this value is used to calculate the dissociation rate for the ScbR homo-dimer formation. Therefore the range of the <math>k^{-}_{3}</math> values is <math>2.12-6.84 min^{-1}</math>, calculated as per <math>k^{-}_{6}=K_{d6} \cdot K_{a}</math>. |
[[Image:K3-text.png|center|thumb|350px|Northrup et al. 1992<ref name="Northrup1992"></ref>]] | [[Image:K3-text.png|center|thumb|350px|Northrup et al. 1992<ref name="Northrup1992"></ref>]] | ||
|} | |} | ||
Revision as of 19:25, 15 October 2015
Two ScbR (R) proteins bind together to form an ScbR homo-dimer (R2).
Contents
Chemical equation
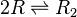
Rate equation
![r= \frac{k^{-}_{6}}{K_{d6}}\cdot [R]^{2} - k^{-}_{6}\cdot [R_{2}]](/wiki/images/math/b/d/2/bd256ae2f0b0944a1e29766d6d3329ee.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of one ScbR to another (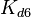 ) and the dissociation rate for binding of one ScbR to another (
) and the dissociation rate for binding of one ScbR to another (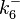 ).
).
| Name | Value | Units | Value in previous GBL models | Remarks-Reference |
|---|---|---|---|---|
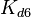
|
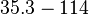 [1] [1]
|

|
N/A | Majka et al. published a study on dimerization of the initiator Protein DnaA of Streptomyces and on its mutants, where they report dissociation constants in the range 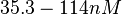 . .
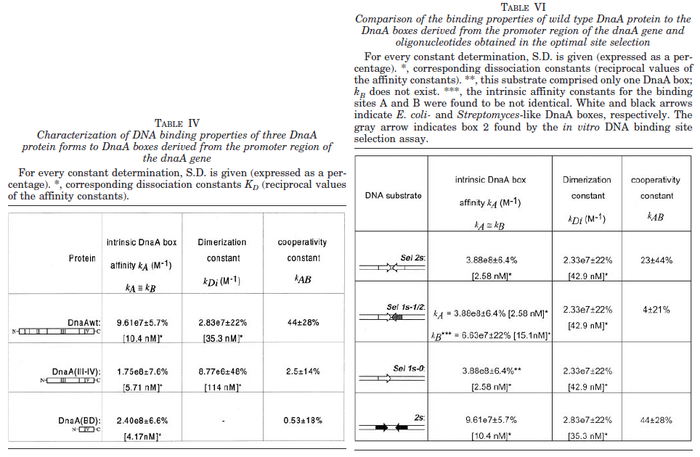 Majka et al. 2001[1] These values agree with Ozbabacan et al. [2] who state that strong protein-protein interactions such as homodimerization have equilibrium dissociation constants |
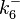
|
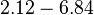 [3] [4] [3] [4]
|

|
N/A | According to Northrup et al. the 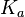 of protein-protein bond formations occur in the order of of protein-protein bond formations occur in the order of 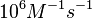 . Therefore, this value is used to calculate the dissociation rate for the ScbR homo-dimer formation. Therefore the range of the . Therefore, this value is used to calculate the dissociation rate for the ScbR homo-dimer formation. Therefore the range of the  values is values is 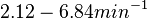 , calculated as per , calculated as per 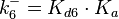 . .
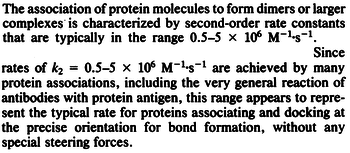 Northrup et al. 1992[4] |
Parameters with uncertainty
The probability distributions, adjusted accordingly in order to reflect the above values, are the following:
The most plausible parameter value for the 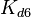 is decided to be
is decided to be 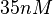 and the confidence interval
and the confidence interval 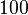 . This means that the mode of the PDF is
. This means that the mode of the PDF is 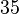 and the range where 95% of the values are found is between
and the range where 95% of the values are found is between 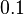 and
and 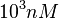 .
.
In a similar way, the most plausible value for 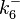 is
is 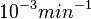 and the confidence interval 10. This means that the mode of the PDF is
and the confidence interval 10. This means that the mode of the PDF is 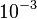 and the range where 95% of the values are found is between
and the range where 95% of the values are found is between 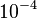 and
and 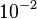
 .
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distributions are:
| Parameter | μ | σ |
|---|---|---|
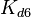
|
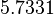
|
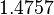
|
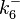
|
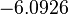
|
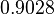
|
References
- ↑ 1.0 1.1 Majka J, Zakrzewska-Czerwiñska J, Messer W. Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J Biol Chem. 2001;276(9):6243-52.
- ↑ Saliha Ece Acuner Ozbabacan, Hatice Billur Engin, Attila Gursoy, and Ozlem Keskin. Transient protein–protein interactions. Protein Engineering, Design and Selection first published online June 15, 2011
- ↑ Janin, Joel. The kinetics of protein-protein recognition. Proteins-Structure Function and Bioinformatics (1997): 153-161.
- ↑ 4.0 4.1 Northrup S.H. and Erickson H.P. Kinetics of protein-protein association explained by Brownian dynamics computer simulation.PNAS 1992;89(8),3338-3342


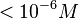 and mostly in the nanomolar range.
and mostly in the nanomolar range.