Difference between revisions of "Formation of homo-dimer R2"
(→Parameters) |
(→Parameters with uncertainty) |
||
| Line 39: | Line 39: | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
| + | The probability distributions, adjusted accordingly in order to reflect the above values, are the following: | ||
| + | |||
| + | The most plausible parameter value for the <math>K_{d6}</math> is decided to be <math> 10 nM </math> and the confidence interval <math> 100 </math>. This means that the mode of the PDF is <math>10</math> and the range where 95% of the values are found is between <math>0.1</math> and <math>10^{3} nM </math>. | ||
| + | |||
| + | In a similar way, the most plausible value for <math>k^{-}_{6}</math> is <math>10^{-3} min^{-1}</math> and the confidence interval 10. This means that the mode of the PDF is <math>10^{-3}</math> and the range where 95% of the values are found is between <math>10^{-4}</math> and <math>10^{-2}</math> <math>min^{-1}</math>. | ||
| + | |||
| + | The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following: | ||
| + | |||
| + | [[Image:Kd6.png|500px]] [[Image:K6.png|500px]] | ||
| + | |||
| + | The location and scale parameters of the distributions are: | ||
| + | {|class="wikitable" | ||
| + | ! Parameter | ||
| + | ! μ | ||
| + | ! σ | ||
| + | |- | ||
| + | |<math>K_{d6}</math> | ||
| + | |<math>4.4804</math> | ||
| + | |<math>1.4757</math> | ||
| + | |- | ||
| + | |<math>k^{-}_{6}</math> | ||
| + | |<math>-6.0926</math> | ||
| + | |<math>0.9028</math> | ||
| + | |} | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 03:07, 5 October 2015
Two ScbR (R) proteins bind together to form an ScbR homo-dimer (R2).
Contents
Chemical equation
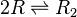
Rate equation
![r= \frac{k^{-}_{6}}{K_{d6}}\cdot [R]^{2} - k^{-}_{6}\cdot [R_{2}]](/wiki/images/math/b/d/2/bd256ae2f0b0944a1e29766d6d3329ee.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of one ScbR to another (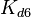 ) and the dissociation rate for binding of one ScbR to another (
) and the dissociation rate for binding of one ScbR to another (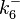 ).
).
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
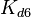
|
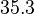 [1] [1]
|

|
Studies with Streptomyces DnaA Protein | |
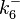
|
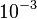 [2] [3] [2] [3]
|

|
Strong protein-protein interactions rate constants |
Parameters with uncertainty
The probability distributions, adjusted accordingly in order to reflect the above values, are the following:
The most plausible parameter value for the 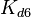 is decided to be
is decided to be 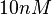 and the confidence interval
and the confidence interval 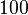 . This means that the mode of the PDF is
. This means that the mode of the PDF is 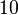 and the range where 95% of the values are found is between
and the range where 95% of the values are found is between 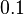 and
and 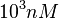 .
.
In a similar way, the most plausible value for 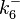 is
is 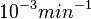 and the confidence interval 10. This means that the mode of the PDF is
and the confidence interval 10. This means that the mode of the PDF is 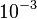 and the range where 95% of the values are found is between
and the range where 95% of the values are found is between 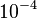 and
and 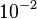
 .
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distributions are:
| Parameter | μ | σ |
|---|---|---|
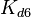
|
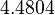
|
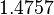
|
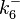
|
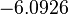
|
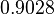
|
References
- ↑ Majka J, Zakrzewska-Czerwiñska J, Messer W. Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J Biol Chem. 2001;276(9):6243-52.
- ↑ Wesley E. Stites. Protein−Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chemical Reviews 1997 97 (5), 1233-1250
- ↑ Janin, Joel. The kinetics of protein-protein recognition. Proteins-Structure Function and Bioinformatics (1997): 153-161.

