Difference between revisions of "Formation of homo-dimer R2"
(→Parameters) |
|||
| Line 31: | Line 31: | ||
|- | |- | ||
|<math>k^{-}_{6}</math> | |<math>k^{-}_{6}</math> | ||
| − | |<math> </math> | + | |<math>10^{-3}</math> <ref name="Stites1997"> Wesley E. Stites. ''Protein−Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis.'' Chemical Reviews 1997 97 (5), 1233-1250</ref> <ref name="Janin1997"> Janin, Joel. ''The kinetics of protein-protein recognition.'' Proteins-Structure Function and Bioinformatics (1997): 153-161.</ref> |
|<math> min^{-1} </math> | |<math> min^{-1} </math> | ||
| − | | | + | | Strong protein-protein interactions rate constants |
| | | | ||
|} | |} | ||
Revision as of 02:50, 5 October 2015
Two ScbR (R) proteins bind together to form an ScbR homo-dimer (R2).
Contents
Chemical equation
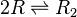
Rate equation
![r= \frac{k^{-}_{6}}{K_{d6}}\cdot [R]^{2} - k^{-}_{6}\cdot [R_{2}]](/wiki/images/math/b/d/2/bd256ae2f0b0944a1e29766d6d3329ee.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of one ScbR to another (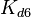 ) and the dissociation rate for binding of one ScbR to another (
) and the dissociation rate for binding of one ScbR to another (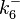 ).
).
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
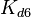
|
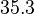 [1] [1]
|

|
Studies with Streptomyces DnaA Protein | |
|
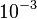 [2] [3] [2] [3]
|

|
Strong protein-protein interactions rate constants |
Parameters with uncertainty
References
- ↑ Majka J, Zakrzewska-Czerwiñska J, Messer W. Sequence recognition, cooperative interaction, and dimerization of the initiator protein DnaA of Streptomyces. J Biol Chem. 2001;276(9):6243-52.
- ↑ Wesley E. Stites. Protein−Protein Interactions: Interface Structure, Binding Thermodynamics, and Mutational Analysis. Chemical Reviews 1997 97 (5), 1233-1250
- ↑ Janin, Joel. The kinetics of protein-protein recognition. Proteins-Structure Function and Bioinformatics (1997): 153-161.

