Difference between revisions of "Diffusion of C and Ce"
(→Parameters) |
(→Parameters) |
||
| (39 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | {{DISPLAYTITLE: Diffusion of C and C<sub>e</sub>}} | ||
The SCB1 protein (C) diffuses from each cell to the environment and back. | The SCB1 protein (C) diffuses from each cell to the environment and back. | ||
<imagemap> | <imagemap> | ||
| Line 11: | Line 12: | ||
== Rate equation == | == Rate equation == | ||
| − | The rate equation is different for the two reactions in order to express the dynamics of the cell density in the environment. Therefore, an additional term <math> | + | The rate equation is different for the two reactions in order to express the dynamics of the cell density in the environment. Therefore, an additional term <math>\rho= V_{cell}/V_{ext}</math> is added in the rate of diffusion from the external environment to the cell. This term represents the additional propensity given to the diffusion of extracellular SCBs by the accumulation of cells in the environment and the consequent increase of cell density. The two rate laws are therefore formed as follows: |
<center><math> r_{out}= D \cdot ([C_{e}]-[C])</math></center> | <center><math> r_{out}= D \cdot ([C_{e}]-[C])</math></center> | ||
| − | <center><math> r_{in}= | + | <center><math> r_{in}= \rho \cdot D \cdot([C]-[C_{e}])</math></center> |
| − | Further information on the coefficient <math> | + | Further information on the coefficient <math>\rho</math> and the dynamics of cell growth and division is provided [[Cellular growth|here]]. |
== Parameters == | == Parameters == | ||
| − | The parameter of this reaction is the diffusion rate of C (<math>D</math>). | + | The parameter of this reaction is the diffusion rate of C (<math>D</math>). In bacteria communication, the autoinducer can either passively diffuse in and out of the cell or, in cases of larger signalling molecules, can be actively transported through the cell membrane. Generally, the diffusion rates when driven by active processes are four orders of magnitude smaller than the passive diffusion rates of small molecules <ref name="Weber2011"> [http://www.biomedcentral.com/content/pdf/1752-0509-5-11.pdf Weber M., Buceta J. ''Noise regulation by quorum sensing in low mRNA copy number systems.'' BMC Systems Biology 2011, 5:11]</ref>. In this model, active transport is not explored, as SCBs are small molecules that can easily diffuse though the cell membrane. However, transport driven by SCB concentration differences due to increase of the cell density in the environment is taken into account. This means that as cell density increases, more autoinducer molecules enter the cell from the outside. The parameter values were derived from measurements and estimations of different autoinducers' passive diffusion and active transport rates in bacteria. |
| − | {|class="wikitable" | + | {|class="wikitable" |
! Name | ! Name | ||
! Value | ! Value | ||
! Units | ! Units | ||
| − | ! | + | ! Value in previous GBL models <ref name="Mehra2008"> [http://www.plosone.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pone.0002724&representation=PDF S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. ''A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor.'' PLoS ONE, 3(7), 2008.] </ref> <ref name="Chatterjee2011"> [http://www.plosone.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pone.0021974&representation=PDF A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. ''Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis.'' PLoS ONE, 6(7), 2011.] </ref> |
| − | ! Remarks | + | ! Remarks-Reference |
|- | |- | ||
|<math>D</math> | |<math>D</math> | ||
| − | |<math>0. | + | |<math>0.1−24</math> <ref name="Kaplan1985"> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC219261/pdf/jbacter00220-0404.pdf Kaplan HB, Greenberg EP. ''Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system''. Journal of Bacteriology. 1985;163(3):1210-1214.]</ref> <ref name="Goryachev2005"> [http://www.ploscompbiol.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pcbi.0010037&representation=PDF Goryachev AB, Toh D-J, Wee KB, Lee T, Zhang H-B, Zhang L-H (2005) ''Transition to Quorum Sensing in an Agrobacterium Population: A Stochastic Model.'' PLoS Comput Biol 1(4): e37.]</ref> <ref name="Groisman2005"> [http://www.nature.com/nmeth/journal/v2/n9/pdf/nmeth784.pdf A. Groisman, C. Lobo, H. Cho, J K. Campbell, Y. S. Dufour, A. M Stevens & A. Levchenko. ''A microfluidic chemostat for experiments with bacterial and yeast cells.'' Nature Methods 2005(2), p. 685 - 689] </ref> |
| − | + | |<math> min^{-1} </math> | |
| − | | | + | |<math>8.3 \cdot 10^{-2} s^{-1} (4.98 min^{-1})</math><ref name="Mehra2008"></ref><ref name="Chatterjee2011"></ref> |
| − | | | + | |
| + | Range tested: <math>0-0.4 s^{-1}</math> | ||
| + | |||
| + | (<math>0-24 min^{-1}</math>) | ||
| + | |||
| + | Bistability range: <math>0.02-0.097 s^{-1}</math><ref name="Mehra2008"></ref> | ||
| + | |||
| + | (<math>1.2-5.82 min^{-1}</math>) | ||
| + | |||
| + | and <math>8.3 \cdot 10^{-6}-4.2 s^{-1}</math><ref name="Chatterjee2011"></ref> | ||
| + | |||
| + | (<math>0.000498-252 min^{-1}</math>) | ||
| + | | Kaplan et al. conducted experiments on how AHL exits and enters the cell in ''Vibrio fischeri'' and reported that the equimolar internal and external concentrations were established within <math>20 s</math> (estimated rate ~<math>3 min^{-1}</math>), suggesting a very fast diffusion rate. If the signal concentration in the medium is sufficiently large, the time for intracellular concentration to reach equilibrium with the extracellular, is <math>~\frac{1}{D}</math>. Therefore, the rate constant of diffusion can be estimated <math>~ \frac{1}{20}s^{-1}=3 min^{-1}</math>. | ||
| + | [[Image:D-text.png|center|thumb|600px|Kaplan et al. 1985<ref name="Kaplan1985"></ref>]] | ||
| + | |||
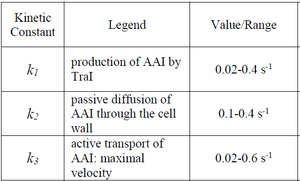
| + | Goryachev et al. have published a stochastic model on quorum sensing in ''Agrobacterium tumefaciens'', where they investigated the role of active transport and passive diffusion of the autoinducer AAI in and out of the cell. The values used for their simulations were <math>0.1-0.4 s^{-1} (6-24 min^{-1})</math> for the passive diffusion and <math>0.02-0.6 s^{-1} (1.2-36 min^{-1})</math> for the active transport. | ||
| + | An estimation within the same range was made by Groisman et al. in a study using a microfluidic chemostat to analyze the cell response to an exogenously added autoinducer, where the reported time span for diffusive exchange of small molecules between different compartments was <math>~40 s</math>. Therefore, the resulting diffusion rate constant is <math>~ \frac{1}{40}s^{-1}=1.5 min^{-1}</math>. | ||
| + | |||
| + | <div><ul> | ||
| + | <li style="display: inline-block;"> [[Image:D-text2.png|center|thumb|300px|[http://journals.plos.org/ploscompbiol/article?id=10.1371/journal.pcbi.0010037#s4 Supplementary material] -Goryachev et al. 2005<ref name="Goryachev2005"></ref>]]</li> | ||
| + | <li style="display: inline-block;"> [[Image:D-text3.png|center|thumb|350px|Groisman et al. 2005<ref name="Groisman2005"></ref>]] </li> | ||
| + | </ul></div> | ||
| + | |||
| + | Finally, Weber et al. reported some cases of active transport where the diffusion rates are much slower, between <math>~0.01</math> and <math>~0.1 min^{-1}</math>. | ||
| + | [[Image:D-text4.png|center|thumb|350px|Weber et al. 2011<ref name="Weber2011"></ref>]] | ||
|} | |} | ||
==Parameters with uncertainty== | ==Parameters with uncertainty== | ||
| − | + | When deciding how to describe the uncertainty for this parameter we must take into consideration that the reported values are either calculated or estimated based on ''in vitro'' and ''in vivo'' experiments in different bacteria species. Additionally, the values correspond to various autoinducers but not to SCBs. This means that there might be a notable difference between actual parameter values and the ones reported in literature. However, the experiments conducted set the general context of diffusion rates for smaller and larger molecules. Therefore, it is safe to assume that GBLs will exhibit a similar behaviour to other small autoinducer molecules. These facts influence the quantification of the parameter uncertainty and therefore the shape of the corresponding distribution. By assigning the appropriate weights to the parameter values and using the method described [[Quantification of parameter uncertainty#Design of probability distributions|'''here''']], the appropriate probability distributions were designed. | |
| + | |||
| + | For this reason, the weight of the distribution is put to <math> 2.8 min^{-1} </math> which is set as the mode of the log-normal distribution for the <math>D</math> and the Spread is <math> 4.27</math>. This means that the range where 68.27% of the values are found is between <math>0.656</math> and <math>11.95 min^{-1}</math>, which also explores a part outside the reported values but also noting the risk that the marginal values would better fit active transport or larger molecules. | ||
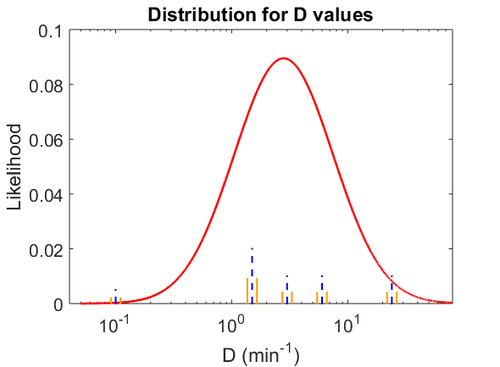
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following: | The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following: | ||
| − | [[Image: | + | [[Image:Du.png|500px]] |
| − | The | + | The values retrieved from literature and their weights are indicated by the blue dashed lines, and the uncertainty for each value is indicated using a default value of 10% error (orange lines). |
| + | |||
| + | The parameter information of the distribution is: | ||
{|class="wikitable" | {|class="wikitable" | ||
! Parameter | ! Parameter | ||
| + | ! Mode | ||
| + | ! Spread | ||
! μ | ! μ | ||
! σ | ! σ | ||
|- | |- | ||
| − | |<math> | + | |<math>D</math> |
| − | |<math>2. | + | |<math>2.8</math> |
| − | |<math>0. | + | |<math>4.27</math> |
| + | |<math>1.9947</math> | ||
| + | |<math>0.98233</math> | ||
|} | |} | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Latest revision as of 18:23, 4 January 2018
The SCB1 protein (C) diffuses from each cell to the environment and back.
Contents
Chemical equation
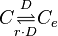
Rate equation
The rate equation is different for the two reactions in order to express the dynamics of the cell density in the environment. Therefore, an additional term 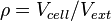 is added in the rate of diffusion from the external environment to the cell. This term represents the additional propensity given to the diffusion of extracellular SCBs by the accumulation of cells in the environment and the consequent increase of cell density. The two rate laws are therefore formed as follows:
is added in the rate of diffusion from the external environment to the cell. This term represents the additional propensity given to the diffusion of extracellular SCBs by the accumulation of cells in the environment and the consequent increase of cell density. The two rate laws are therefore formed as follows:
![r_{out}= D \cdot ([C_{e}]-[C])](/wiki/images/math/b/e/1/be1bfbe3de85b2144bfc73b5cd16a5c2.png)
![r_{in}= \rho \cdot D \cdot([C]-[C_{e}])](/wiki/images/math/9/6/a/96a4cb5ee35a5a2d5f16eb4a39970485.png)
Further information on the coefficient 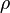 and the dynamics of cell growth and division is provided here.
and the dynamics of cell growth and division is provided here.
Parameters
The parameter of this reaction is the diffusion rate of C (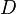 ). In bacteria communication, the autoinducer can either passively diffuse in and out of the cell or, in cases of larger signalling molecules, can be actively transported through the cell membrane. Generally, the diffusion rates when driven by active processes are four orders of magnitude smaller than the passive diffusion rates of small molecules [1]. In this model, active transport is not explored, as SCBs are small molecules that can easily diffuse though the cell membrane. However, transport driven by SCB concentration differences due to increase of the cell density in the environment is taken into account. This means that as cell density increases, more autoinducer molecules enter the cell from the outside. The parameter values were derived from measurements and estimations of different autoinducers' passive diffusion and active transport rates in bacteria.
). In bacteria communication, the autoinducer can either passively diffuse in and out of the cell or, in cases of larger signalling molecules, can be actively transported through the cell membrane. Generally, the diffusion rates when driven by active processes are four orders of magnitude smaller than the passive diffusion rates of small molecules [1]. In this model, active transport is not explored, as SCBs are small molecules that can easily diffuse though the cell membrane. However, transport driven by SCB concentration differences due to increase of the cell density in the environment is taken into account. This means that as cell density increases, more autoinducer molecules enter the cell from the outside. The parameter values were derived from measurements and estimations of different autoinducers' passive diffusion and active transport rates in bacteria.
| Name | Value | Units | Value in previous GBL models [2] [3] | Remarks-Reference |
|---|---|---|---|---|
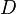
|
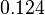 [4] [5] [6] [4] [5] [6]
|

|
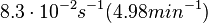 [2][3] [2][3]
Range tested: ( Bistability range: ( and ( |
Kaplan et al. conducted experiments on how AHL exits and enters the cell in Vibrio fischeri and reported that the equimolar internal and external concentrations were established within 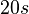 (estimated rate ~ (estimated rate ~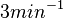 ), suggesting a very fast diffusion rate. If the signal concentration in the medium is sufficiently large, the time for intracellular concentration to reach equilibrium with the extracellular, is ), suggesting a very fast diffusion rate. If the signal concentration in the medium is sufficiently large, the time for intracellular concentration to reach equilibrium with the extracellular, is 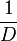 . Therefore, the rate constant of diffusion can be estimated . Therefore, the rate constant of diffusion can be estimated 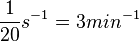 . .
 Kaplan et al. 1985[4] Goryachev et al. have published a stochastic model on quorum sensing in Agrobacterium tumefaciens, where they investigated the role of active transport and passive diffusion of the autoinducer AAI in and out of the cell. The values used for their simulations were
Finally, Weber et al. reported some cases of active transport where the diffusion rates are much slower, between  Weber et al. 2011[1] |
Parameters with uncertainty
When deciding how to describe the uncertainty for this parameter we must take into consideration that the reported values are either calculated or estimated based on in vitro and in vivo experiments in different bacteria species. Additionally, the values correspond to various autoinducers but not to SCBs. This means that there might be a notable difference between actual parameter values and the ones reported in literature. However, the experiments conducted set the general context of diffusion rates for smaller and larger molecules. Therefore, it is safe to assume that GBLs will exhibit a similar behaviour to other small autoinducer molecules. These facts influence the quantification of the parameter uncertainty and therefore the shape of the corresponding distribution. By assigning the appropriate weights to the parameter values and using the method described here, the appropriate probability distributions were designed.
For this reason, the weight of the distribution is put to 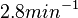 which is set as the mode of the log-normal distribution for the
which is set as the mode of the log-normal distribution for the 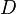 and the Spread is
and the Spread is 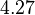 . This means that the range where 68.27% of the values are found is between
. This means that the range where 68.27% of the values are found is between 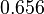 and
and 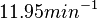 , which also explores a part outside the reported values but also noting the risk that the marginal values would better fit active transport or larger molecules.
, which also explores a part outside the reported values but also noting the risk that the marginal values would better fit active transport or larger molecules.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The values retrieved from literature and their weights are indicated by the blue dashed lines, and the uncertainty for each value is indicated using a default value of 10% error (orange lines).
The parameter information of the distribution is:
| Parameter | Mode | Spread | μ | σ |
|---|---|---|---|---|
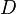
|
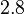
|
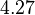
|
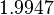
|
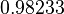
|
References
- ↑ 1.0 1.1 Weber M., Buceta J. Noise regulation by quorum sensing in low mRNA copy number systems. BMC Systems Biology 2011, 5:11
- ↑ 2.0 2.1 2.2 S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 3.0 3.1 3.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ 4.0 4.1 Kaplan HB, Greenberg EP. Diffusion of autoinducer is involved in regulation of the Vibrio fischeri luminescence system. Journal of Bacteriology. 1985;163(3):1210-1214.
- ↑ 5.0 5.1 Goryachev AB, Toh D-J, Wee KB, Lee T, Zhang H-B, Zhang L-H (2005) Transition to Quorum Sensing in an Agrobacterium Population: A Stochastic Model. PLoS Comput Biol 1(4): e37.
- ↑ 6.0 6.1 A. Groisman, C. Lobo, H. Cho, J K. Campbell, Y. S. Dufour, A. M Stevens & A. Levchenko. A microfluidic chemostat for experiments with bacterial and yeast cells. Nature Methods 2005(2), p. 685 - 689


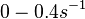
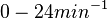 )
)
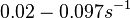
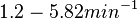 )
)
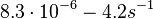
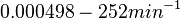 )
)
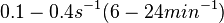 for the passive diffusion and
for the passive diffusion and 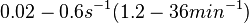 for the active transport.
An estimation within the same range was made by Groisman et al. in a study using a microfluidic chemostat to analyze the cell response to an exogenously added autoinducer, where the reported time span for diffusive exchange of small molecules between different compartments was
for the active transport.
An estimation within the same range was made by Groisman et al. in a study using a microfluidic chemostat to analyze the cell response to an exogenously added autoinducer, where the reported time span for diffusive exchange of small molecules between different compartments was 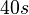 . Therefore, the resulting diffusion rate constant is
. Therefore, the resulting diffusion rate constant is 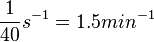 .
.

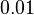 and
and 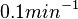 .
.