Difference between revisions of "Degradation of r-a"
(→Parameters) |
|||
| Line 17: | Line 17: | ||
== Parameters == | == Parameters == | ||
| − | The parameter of this reaction is the degradation rate of the r-a complex <math> | + | The parameter of this reaction is the degradation rate of the r-a complex <math>d_{mRA}</math>. |
| − | {|class="wikitable" | + | {|class="wikitable" |
! Name | ! Name | ||
! Value | ! Value | ||
! Units | ! Units | ||
| − | ! | + | ! Value in previous GBL model <ref name="Chatterjee2011"> [http://www.plosone.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pone.0021974&representation=PDF A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. ''Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis.'' PLoS ONE, 6(7), 2011.] </ref> |
| − | ! Remarks | + | ! Remarks-Reference |
|- | |- | ||
| − | |<math>d_{mRA} </math> | + | |<math>d_{mRA}</math> |
| − | |<math>0. | + | |<math>0.03-0.4</math> <ref name="Carpousis2003"> [http://genesdev.cshlp.org/content/17/19/2351.full.pdf Carpousis A.J. ''Degradation of targeted mRNAs in Escherichia coli: regulation by a small antisense RNA''. Genes & Dev. 2003. 17: 2351-2355.]</ref> <ref name="Georg2011"> [http://mmbr.asm.org/content/75/2/286.full.pdf Georg J., Hess WR. ''Cis-antisense RNA, another level of gene regulation in bacteria''. Microbiol Mol Biol Rev. 2011;75(2):286-300.]</ref> |
| − | |<math>min^{-1} </math> | + | |<math> min^{-1} </math> |
| − | | | + | |<math>0.01 s^{-1}</math><ref name="Chatterjee2011"></ref> |
| − | | | + | |
| + | (Bistability range: and <math>0-1.24 s^{-1}) | ||
| + | | In a study on the properties of antisense RNA and on the effect it has on gene regulation in bacteria, Georg et al. report that the antisense RNAs resulting from overlapping or divergent promoters (i.e. ''isiA''/IsrR sense/antisense pair in ''Synechocystis'' PCC6803) that form a complex, co-degrade almost immediately. | ||
| + | [[Image:DmRA-text.png|center|thumb|600px|Georg et al. 2012<ref name="Georg2011"></ref>]] | ||
| + | |||
| + | This opinion is further supported by A.J. Carpousis who, in a review on gene regulation by antisense RNA and quantifies a short mRNA half-life as approximately <math>2 min</math>. | ||
| + | [[Image:DmRA-text2.png|center|thumb|500px|A.J. Carpousis 2003<ref name="Carpousis2003"></ref>]] | ||
| + | |||
| + | Therefore we can assume that the antisense RNA in the GBL system will follow a similar pattern and will rapidly degrade. The half-life should be kept under <math>2 min</math>, therefore we can explore the range of values <math>0.01-2 min</math>. From the antisense RNA half-life (<math>t_{1/2}</math>) we calculated the degradation rate, as per <math>d_{mRA}= \frac{ln(2)}{t_{1/2}}</math>, which resulted in a degradation rate constant value between <math>0.35-69.3 min^{-1} </math>. | ||
|} | |} | ||
Revision as of 23:39, 16 October 2015
[NOTE TO MARC: Can you check the parameter for this reaction? I assumed the mRNA complex half life to be 1 min, is it too slow?]
The mRNA complex of scbR-scbA degrades very soon after it's formation.
Contents
Chemical equation
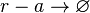
Rate equation
![r= d_{mAR}\cdot[r-a]](/wiki/images/math/d/9/9/d99d29699d17e05bf32ad10bd105c21b.png)
Parameters
The parameter of this reaction is the degradation rate of the r-a complex 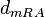 .
.
| Name | Value | Units | Value in previous GBL model [1] | Remarks-Reference |
|---|---|---|---|---|
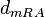
|
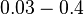 [2] [3] [2] [3]
|

|
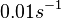 [1] [1]
(Bistability range: and Failed to parse (lexing error): 0-1.24 s^{-1}) | In a study on the properties of antisense RNA and on the effect it has on gene regulation in bacteria, Georg et al. report that the antisense RNAs resulting from overlapping or divergent promoters (i.e. ''isiA''/IsrR sense/antisense pair in ''Synechocystis'' PCC6803) that form a complex, co-degrade almost immediately. [[Image:DmRA-text.png|center|thumb|600px|Georg et al. 2012<ref name="Georg2011"></ref>]] This opinion is further supported by A.J. Carpousis who, in a review on gene regulation by antisense RNA and quantifies a short mRNA half-life as approximately <math>2 min .  A.J. Carpousis 2003[2] Therefore we can assume that the antisense RNA in the GBL system will follow a similar pattern and will rapidly degrade. The half-life should be kept under |
Parameters with uncertainty
References
- ↑ 1.0 1.1 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ 2.0 2.1 Carpousis A.J. Degradation of targeted mRNAs in Escherichia coli: regulation by a small antisense RNA. Genes & Dev. 2003. 17: 2351-2355.
- ↑ Georg J., Hess WR. Cis-antisense RNA, another level of gene regulation in bacteria. Microbiol Mol Biol Rev. 2011;75(2):286-300.


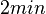 , therefore we can explore the range of values
, therefore we can explore the range of values 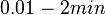 . From the antisense RNA half-life (
. From the antisense RNA half-life (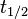 ) we calculated the degradation rate, as per
) we calculated the degradation rate, as per 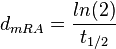 , which resulted in a degradation rate constant value between
, which resulted in a degradation rate constant value between 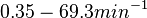 .
.