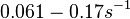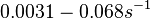Degradation of C2-R2
The SCB-ScbR protein complex (C2R2) degrades.
Contents
Chemical equation
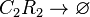
Rate equation
![r= d_{CR}\cdot[C_{2}R_{2}]](/wiki/images/math/f/a/d/fad2befe488ac08f5d5d5486e955d97e.png)
Parameters
The parameter of this reaction is the degradation rate of C2R2 (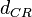 ). The parameter values were derived from proteomics studies on different proteins of S. coelicolor.
). The parameter values were derived from proteomics studies on different proteins of S. coelicolor.
| Name | Value | Units | Value in previous GBL models [1] [2] | Remarks-Reference |
|---|---|---|---|---|
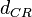
|
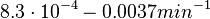 [3] [4] [3] [4]
|

|
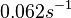 [1] [1]
(Range tested: (Bistability range: and |
In a quantitative proteomics study on protein turnover rates in dynamic systems, Jayapal et al. reported the degradation rates of 115 proteins in S. coelicolor cultures undergoing transition from exponential growth to stationary phase. The values were in the range 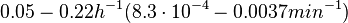 with a median of with a median of 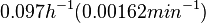 . .
 Jayapal et al. 2012[4] The full list of the proteins studied and their turnover rate constants can be found here. |
Parameters with uncertainty
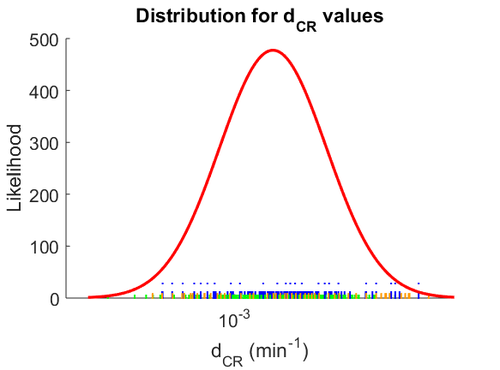
When deciding how to describe the uncertainty for this parameter we must take into consideration that the reported values are estimated or calculated from proteomics experiments, using methods which were prone to measurement errors (i.e. optical density of fragmented mycelia to estimate the growth rate (μ)) and extrapolations from cell density to total cellular protein without taking into account the dilution due to cell division. Additionally, the study did not include measurements on protein complexes, although it was conducted on a wide range of S. coelicolor proteins which enables a rough estimation of the range of values of protein degradation in this species. These facts influence the quantification of the parameter uncertainty and therefore the shape of the corresponding distribution.
Therefore, although the weight of the distribution is put on the reported range of values by setting the mode of the log-normal distribution of the 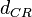 to
to 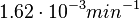 which was the reported median value in the study, we wish to explore the full range of published values and even sample a percentage of values outside the reported range. For this reason, the confidence interval factor is set to
which was the reported median value in the study, we wish to explore the full range of published values and even sample a percentage of values outside the reported range. For this reason, the confidence interval factor is set to 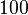 , so that the range where 95% of the values are found is between
, so that the range where 95% of the values are found is between 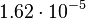 and
and 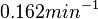 .
.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
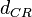
|
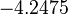
|
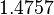
|
References
- ↑ 1.0 1.1 1.2 S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 2.0 2.1 2.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ Trötschel C, Albaum SP, Poetsch A. Proteome turnover in bacteria: current status for Corynebacterium glutamicum and related bacteria. Microbial biotechnology. 2013;6(6):708-719.
- ↑ 4.0 4.1 Jayapal KP, Sui S, Philp RJ, Kok YJ, Yap MG, Griffin TJ, Hu WS. Multitagging proteomic strategy to estimate protein turnover rates in dynamic systems. J Proteome Res. 2010 May 7;9(5):2087-97.


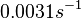
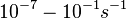 )
)