Degradation of C
The SCB protein (C) degrades.
Contents
Chemical equation
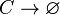
Rate equation
![r= d_{C}\cdot[C]](/wiki/images/math/f/c/b/fcb71209c990f6af04af87f99fed6a0f.png)
Parameters
The parameter of this reaction is the degradation rate of C (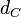 ).
).
| Name | Value | Units | Value in previous GBL models [1] [2] | Remarks-Reference |
|---|---|---|---|---|
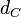
|
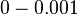 [3] [4] [3] [4]
|
 [5] [6] [5] [6]
|
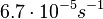 [1][2] [1][2]
(Range tested: (Bistability range: and |
Chen et al. empirically calculated the degradation rate of the autoinducer PAI2 in Pseudomonas aeruginosa cultures and reported a best-fit degradation constant of 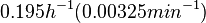 . .
 Chen et al. 2005[4] Additionally, the degradation rates of the quorum sensing autoinducer AHL have been measured in vitro by Kaufmann et al. and the reported rates for different AHLs are between |
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
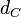
|
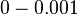 [7] [7]
|

|
Degradation rate of acyl-homoserine lactone (AHL) | The assumption that the degradation of SCB1 is slower than
AHL is made, as described in Takano(2006)[3] |
Parameters with uncertainty
The most plausible parameter value for the 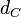 is decided to be
is decided to be 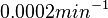 and the confidence interval
and the confidence interval 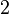 . This means that the mode of the PDF is 0.0002 and the range where 95% of the values are found is between
. This means that the mode of the PDF is 0.0002 and the range where 95% of the values are found is between 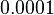 and
and 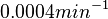 .
.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
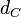
|
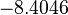
|
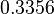
|
References
- ↑ 1.0 1.1 1.2 S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 2.0 2.1 2.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ 3.0 3.1 E. Takano. γ-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Current Opinion in Microbiology, 9(3):287–294, 2006. Cite error: Invalid
<ref>tag; name "Takano2006" defined multiple times with different content - ↑ 4.0 4.1 Chen CC., Riadi L., Suh S.J., Ohman D.E., Ju L.K. Degradation and synthesis kinetics of quorum-sensing autoinducer in Pseudomonas aeruginosa cultivation. J Biotechnol. 2005;117(1):1-10.
- ↑ 5.0 5.1 Weber M., Buceta J. Noise regulation by quorum sensing in low mRNA copy number systems. BMC Systems Biology 2011, 5:11
- ↑ 6.0 6.1 Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. Revisiting quorum sensing: Discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci U S A. 2005;102(2):309-14.
- ↑ S. Basu, Y. Gerchman, C. H. Collins, F. H. Arnold & R. Weiss. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130-1134, 2005


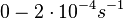 )
)
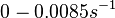
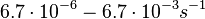
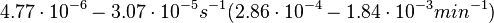 . Weber et al. also reported that the degradation rate of AHL in vivo has been estimated, and is within the range
. Weber et al. also reported that the degradation rate of AHL in vivo has been estimated, and is within the range 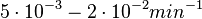 .
.

