Difference between revisions of "Degradation of C"
(→Parameters) |
(→Parameters) |
||
| Line 15: | Line 15: | ||
== Parameters == | == Parameters == | ||
| − | The parameter of this reaction is the degradation rate of C (<math>d_{C}</math>). | + | The parameter of this reaction is the degradation rate of C (<math>d_{C}</math>). The parameter values were derived from measurements and estimations of autoinducer degradation rates from other bacteria (AHL, PAI2). |
{|class="wikitable" | {|class="wikitable" | ||
| Line 43: | Line 43: | ||
</ul></div> | </ul></div> | ||
| − | + | With regards to SCBs in particular, according to a review by E. Takano these molecules are very stable and can be still detected after more than 12h into stationary phase. | |
| − | + | [[Image:DC-text4.png|center|thumb|350px|E. Takano 2006<ref name="Takano2006"></ref>]] | |
| − | + | Therefore, we can assume that the degradation rate of SCB is slower than the ones reported for other autoinducers and will not exceed the value <math>2 \cdot 10^{-2} min^{-1}</math>. | |
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | | | ||
| − | | | ||
| − | | | ||
| − | |||
| − | |||
| − | |||
| − | |||
|} | |} | ||
Revision as of 19:19, 17 October 2015
The SCB protein (C) degrades.
Contents
Chemical equation
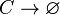
Rate equation
![r= d_{C}\cdot[C]](/wiki/images/math/f/c/b/fcb71209c990f6af04af87f99fed6a0f.png)
Parameters
The parameter of this reaction is the degradation rate of C (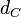 ). The parameter values were derived from measurements and estimations of autoinducer degradation rates from other bacteria (AHL, PAI2).
). The parameter values were derived from measurements and estimations of autoinducer degradation rates from other bacteria (AHL, PAI2).
| Name | Value | Units | Value in previous GBL models [1] [2] | Remarks-Reference |
|---|---|---|---|---|
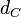
|
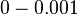 [3] [4] [3] [4]
|
 [5] [6] [5] [6]
|
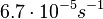 [1][2] [1][2]
(Range tested: (Bistability range: and |
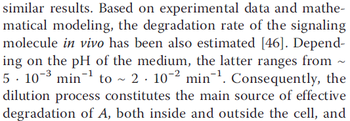
Chen et al. empirically calculated the degradation rate of the autoinducer PAI2 in Pseudomonas aeruginosa cultures and reported a best-fit degradation constant of 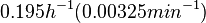 . .
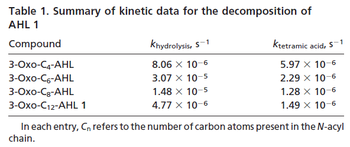
 Chen et al. 2005[4] Additionally, the degradation rates of the quorum sensing autoinducer AHL have been measured in vitro by Kaufmann et al. and the reported rates for different AHLs are between With regards to SCBs in particular, according to a review by E. Takano these molecules are very stable and can be still detected after more than 12h into stationary phase.  E. Takano 2006[3] Therefore, we can assume that the degradation rate of SCB is slower than the ones reported for other autoinducers and will not exceed the value |
Parameters with uncertainty
The most plausible parameter value for the 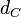 is decided to be
is decided to be 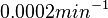 and the confidence interval
and the confidence interval 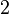 . This means that the mode of the PDF is 0.0002 and the range where 95% of the values are found is between
. This means that the mode of the PDF is 0.0002 and the range where 95% of the values are found is between 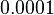 and
and 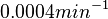 .
.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
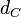
|
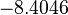
|
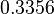
|
References
- ↑ 1.0 1.1 1.2 S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 2.0 2.1 2.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ 3.0 3.1 E. Takano. γ-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Current Opinion in Microbiology, 9(3):287–294, 2006.
- ↑ 4.0 4.1 Chen CC., Riadi L., Suh S.J., Ohman D.E., Ju L.K. Degradation and synthesis kinetics of quorum-sensing autoinducer in Pseudomonas aeruginosa cultivation. J Biotechnol. 2005;117(1):1-10.
- ↑ 5.0 5.1 Weber M., Buceta J. Noise regulation by quorum sensing in low mRNA copy number systems. BMC Systems Biology 2011, 5:11
- ↑ 6.0 6.1 Kaufmann GF, Sartorio R, Lee SH, Rogers CJ, Meijler MM, Moss JA, Clapham B, Brogan AP, Dickerson TJ, Janda KD. Revisiting quorum sensing: Discovery of additional chemical and biological functions for 3-oxo-N-acylhomoserine lactones. Proc Natl Acad Sci U S A. 2005;102(2):309-14.


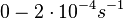 )
)
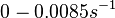
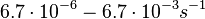
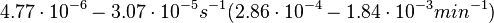 . Weber et al. also reported that the degradation rate of AHL in vivo has been estimated, and is within the range
. Weber et al. also reported that the degradation rate of AHL in vivo has been estimated, and is within the range 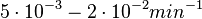 .
.
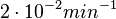 .
.