Difference between revisions of "Degradation of C"
(→Parameters) |
|||
| Line 16: | Line 16: | ||
The parameter of this reaction is the degradation rate of C (<math>d_{C}</math>). | The parameter of this reaction is the degradation rate of C (<math>d_{C}</math>). | ||
| + | |||
| + | {|class="wikitable" | ||
| + | ! Name | ||
| + | ! Value | ||
| + | ! Units | ||
| + | ! Value in previous GBL models <ref name="Mehra2008"> [http://www.plosone.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pone.0002724&representation=PDF S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. ''A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor.'' PLoS ONE, 3(7), 2008.] </ref> <ref name="Chatterjee2011"> [http://www.plosone.org/article/fetchObject.action?uri=info:doi/10.1371/journal.pone.0021974&representation=PDF A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. ''Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis.'' PLoS ONE, 6(7), 2011.] </ref> | ||
| + | ! Remarks-Reference | ||
| + | |- | ||
| + | |<math>d_{C}</math> | ||
| + | |<math>0-0.001</math> <ref name="Kristoffersen2012"> [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3446304/pdf/gb-2012-13-4-r30.pdf Kristoffersen SM, Haase C, Weil MR, et al. ''Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium.'' Genome Biology. 2012;13(4):R30.]</ref> <ref name="Ortiz1984"> [http://www.sciencedirect.com/science/book/9780125286206 Ortiz-Ortiz, L. and Bojalil, L.F. and Yakoleff, V. ''Biological, Biochemical, and Biomedical Aspects of Actinomycetes.'' Elsevier, 1984]</ref> | ||
| + | |<math> min^{-1} </math> | ||
| + | |<math>6.7 \cdot 10^{-5} s^{-1}</math><ref name="Mehra2008"></ref><ref name="Chatterjee2011"></ref> | ||
| + | |||
| + | (Range tested: <math>0-2 \cdot 10^{-4} s^{-1}</math>) | ||
| + | |||
| + | (Bistability range: <math>0-0.0085 s^{-1}</math><ref name="Mehra2008"></ref> | ||
| + | |||
| + | and <math>6.7 \cdot 10^{-6}-6.7 \cdot 10^{-3} s^{-1}</math><ref name="Chatterjee2011"></ref>) | ||
| + | | In a study on global analysis of mRNA decay in gram positive bacteria by RNA-Seq, Kristoffersen et al. reported a range of values for the half-life (<math>t_{1/2}</math>) of mRNA in ''B. cereus'' between <math>1.9-5.6 min </math>, with a median of <math>2.4 min </math>. From these values we calculated the mRNA degradation rate of ''scbR'' as per <math>d_{mR}= \frac{ln(2)}{t_{1/2}}</math>, which resulted in a degradation rate constant value range <math>0.124-0.365 min^{-1} </math> and a median <math>0.29 min^{-1} </math>. | ||
| + | [[Image:DmR-text.png|center|thumb|600px|Kristoffersen et al. 2012<ref name="Kristoffersen2012"></ref>]] | ||
| + | |||
| + | Additionally, in a book on biological and biochemical aspects of ''Actinomycetes'', Ortiz-Ortiz et al. reported a range of mRNA degradation rates for ''Streptomyces antibioticus'' spores between <math>4-22.5 min </math>. By the same calculations, the corresponding mRNA degradation rate constants are in the range <math> 0.031-0.17 min^{-1} </math>. | ||
| + | [[Image:DmR-text2.png|center|thumb|500px|Ortiz-Ortiz et al. 1984<ref name="Ortiz1984"></ref>]] | ||
| + | |} | ||
{|class="wikitable" | {|class="wikitable" | ||
Revision as of 04:35, 17 October 2015
The SCB protein (C) degrades.
Contents
Chemical equation
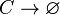
Rate equation
![r= d_{C}\cdot[C]](/wiki/images/math/f/c/b/fcb71209c990f6af04af87f99fed6a0f.png)
Parameters
The parameter of this reaction is the degradation rate of C (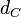 ).
).
| Name | Value | Units | Value in previous GBL models [1] [2] | Remarks-Reference |
|---|---|---|---|---|
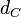
|
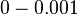 [3] [4] [3] [4]
|

|
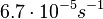 [1][2] [1][2]
(Range tested: (Bistability range: and |
In a study on global analysis of mRNA decay in gram positive bacteria by RNA-Seq, Kristoffersen et al. reported a range of values for the half-life (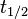 ) of mRNA in B. cereus between ) of mRNA in B. cereus between 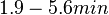 , with a median of , with a median of 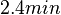 . From these values we calculated the mRNA degradation rate of scbR as per . From these values we calculated the mRNA degradation rate of scbR as per 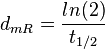 , which resulted in a degradation rate constant value range , which resulted in a degradation rate constant value range 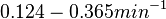 and a median and a median 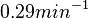 . .
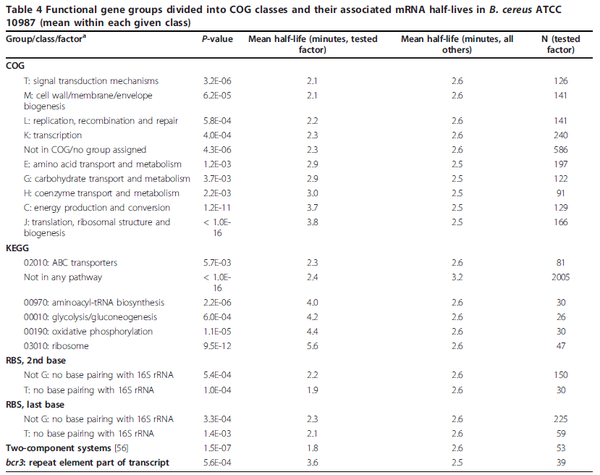 Kristoffersen et al. 2012[3] Additionally, in a book on biological and biochemical aspects of Actinomycetes, Ortiz-Ortiz et al. reported a range of mRNA degradation rates for Streptomyces antibioticus spores between 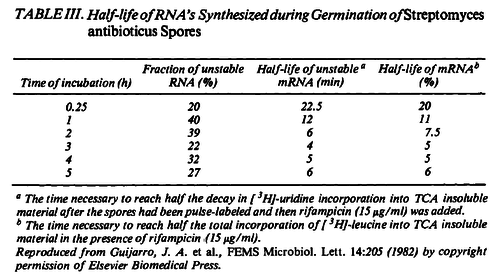 Ortiz-Ortiz et al. 1984[4] |
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
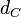
|
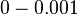 [5] [5]
|

|
Degradation rate of acyl-homoserine lactone (AHL) | The assumption that the degradation of SCB1 is slower than
AHL is made, as described in Takano(2006)[6] |
Parameters with uncertainty
The most plausible parameter value for the 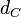 is decided to be
is decided to be 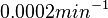 and the confidence interval
and the confidence interval 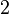 . This means that the mode of the PDF is 0.0002 and the range where 95% of the values are found is between
. This means that the mode of the PDF is 0.0002 and the range where 95% of the values are found is between 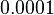 and
and 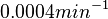 .
.
The probability distribution for the parameter, adjusted accordingly in order to reflect the above values, is the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
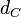
|
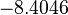
|
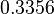
|
References
- ↑ 1.0 1.1 1.2 S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ 2.0 2.1 2.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ 3.0 3.1 Kristoffersen SM, Haase C, Weil MR, et al. Global mRNA decay analysis at single nucleotide resolution reveals segmental and positional degradation patterns in a Gram-positive bacterium. Genome Biology. 2012;13(4):R30.
- ↑ 4.0 4.1 Ortiz-Ortiz, L. and Bojalil, L.F. and Yakoleff, V. Biological, Biochemical, and Biomedical Aspects of Actinomycetes. Elsevier, 1984
- ↑ S. Basu, Y. Gerchman, C. H. Collins, F. H. Arnold & R. Weiss. A synthetic multicellular system for programmed pattern formation. Nature 434, 1130-1134, 2005
- ↑ E. Takano. γ-butyrolactones: Streptomyces signalling molecules regulating antibiotic production and differentiation. Current Opinion in Microbiology, 9(3):287–294, 2006.


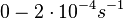 )
)
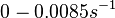
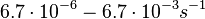
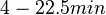 . By the same calculations, the corresponding mRNA degradation rate constants are in the range
. By the same calculations, the corresponding mRNA degradation rate constants are in the range 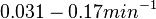 .
.