Binding of R2 to OA operator
The ScbR homo-dimer (R2) binds to the ScbA gene promoter (OA) and represses its mRNA transcription.
Contents
Chemical equation
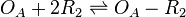
Rate equation
![r= \frac{k^{-}_{2}}{K_{d2}}\cdot [O_{A}]\cdot [R_{2}]^{2} - k^{-}_{2}\cdot [O_{A}-R_{2}]](/wiki/images/math/4/8/5/48594eae1121e765c17117c9d77e08da.png)
Parameters
The parameters of this reaction are the dissociation constant for binding of ScbR to OA (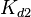 ) and the dissociation rate for binding of ScbR to OA (
) and the dissociation rate for binding of ScbR to OA (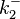 ).
).
| Name | Value | Units | Origin | Remarks |
|---|---|---|---|---|
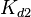
|
 [1] [2] [3] [1] [2] [3]
|

|
TetR-tetO interaction and
TetR-like Rv3066 from M. tuberculosis |
Repressor protein TetR binds to the operator tetO, repressing its own expression and that of the
efflux determinant tetA. Similar structure and activity as ScbR binding to OA |
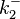
|
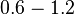 [4] [5] [4] [5]
|

|
SPR of a TetR-like protein (RolR) on a Gram and
GC content ~ 50-60% from Corynebacterium glutamicum |
Parameters with uncertainty
The most plausible parameter value for the 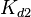 is decided to be
is decided to be 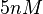 and the confidence interval
and the confidence interval 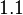 . This means that the mode of the PDF is 5 and the range where 95% of the values are found is between 4.55 and 5.5 nM.
. This means that the mode of the PDF is 5 and the range where 95% of the values are found is between 4.55 and 5.5 nM.
In a similar way, the most plausible value for 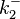 is
is 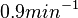 and the confidence interval
and the confidence interval 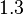 . This means that the mode of the PDF is 0.9 and the range where 95% of the values are found is between 0.6923 and 1.17
. This means that the mode of the PDF is 0.9 and the range where 95% of the values are found is between 0.6923 and 1.17  .
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distributions are:
| Parameter | μ | σ |
|---|---|---|
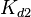
|
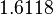
|
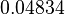
|
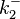
|
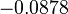
|
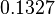
|
References
- ↑ Kamionka A, Bogdanska-Urbaniak J, Scholz O, Hillen W. Two mutations in the tetracycline repressor change the inducer anhydrotetracycline to a corepressor. Nucleic Acids Research. 2004;32(2):842-847.
- ↑ Bolla JR, Do SV, Long F, et al. Structural and functional analysis of the transcriptional regulator Rv3066 of Mycobacterium tuberculosis. Nucleic Acids Research. 2012;40(18):9340-9355.
- ↑ Ahn SK, Tahlan K, Yu Z, Nodwell J. Investigation of Transcription Repression and Small-Molecule Responsiveness by TetR-Like Transcription Factors Using a Heterologous Escherichia coli-Based Assay. Journal of Bacteriology. 2007;189(18):6655-6664.
- ↑ Sylwia Kedracka-Krok, Andrzej Gorecki, Piotr Bonarek, and Zygmunt Wasylewski. Kinetic and Thermodynamic Studies of Tet Repressor−Tetracycline Interaction. Biochemistry 2005 44 (3), 1037-1046.
- ↑ Li T, Zhao K, Huang Y, et al. The TetR-Type Transcriptional Repressor RolR from Corynebacterium glutamicum Regulates Resorcinol Catabolism by Binding to a Unique Operator, rolO. Applied and Environmental Microbiology. 2012;78(17):6009-6016.

