Difference between revisions of "Background Information on GBL system"
(→Regulation in Streptomyces coelicolor) |
(→Regulation in Streptomyces coelicolor) |
||
| Line 16: | Line 16: | ||
Recent studies and reviews \citep{VanWezel20111311,Liu2013112} point to the hypothesis that the GBL regulatory system involves a small, yet complex two gene network composed by a synthase (''scbA'' (SCO6266)) and a butyrolactone receptor (''scbR'' (SCO6265)), which governs a potentially bistable switch between the "on" and "off" states of antibiotic production. The ''scbA'' gene product is presumed to catalyse the condensation of dihydroxyacetone phosphate with a beta--ketoacid to produce three different butyrolactones (SCB1, SCB2 and SCB3). \citep{Takano2006287,kato2007biosynthesis} On the other hand, ''scbR'' is a TetR-like \citep{Ramos2005326} DNA-binding protein known to regulate its own transcription and that of ''scbA''. Additionally, ScbR protein has a γ-butyrolactone binding domain at its C-terminal and a DNA binding domain at its N-terminal \citep{natsume2004crystal}. This structure allows it to directly regulate the production of a cryptic metabolite and indirectly the production of blue pigmented actinorhodin (Act) and prodigiosins (e.g. undecylprodigiosin, Red). These genes are divergently encoded and their promoter regions overlap 53bp, as has been shown by gel retardation assays and DNase I footprinting studies. \citep{takano2001complex} Complementary to these findings, the discovery of cis asRNA has also been reported in different studies. \citep{d2010noncoding,panek2008biocomputational,swiercz2008small} The regulatory role of the system's overlapping topology and of the antisense RNA in biological decision making has been demonstrated in number of studies concerning different bacteria, such as ''E. faecalis'' \citep{chatterjee2011convergent} and ''S. enterica''. \citep{lee2010antisense} It is therefore possible that the formation of an antisense RNA between the ''scbA'' and ''scbR'' transcripts could play an important role in the regulation of the GBL system. | Recent studies and reviews \citep{VanWezel20111311,Liu2013112} point to the hypothesis that the GBL regulatory system involves a small, yet complex two gene network composed by a synthase (''scbA'' (SCO6266)) and a butyrolactone receptor (''scbR'' (SCO6265)), which governs a potentially bistable switch between the "on" and "off" states of antibiotic production. The ''scbA'' gene product is presumed to catalyse the condensation of dihydroxyacetone phosphate with a beta--ketoacid to produce three different butyrolactones (SCB1, SCB2 and SCB3). \citep{Takano2006287,kato2007biosynthesis} On the other hand, ''scbR'' is a TetR-like \citep{Ramos2005326} DNA-binding protein known to regulate its own transcription and that of ''scbA''. Additionally, ScbR protein has a γ-butyrolactone binding domain at its C-terminal and a DNA binding domain at its N-terminal \citep{natsume2004crystal}. This structure allows it to directly regulate the production of a cryptic metabolite and indirectly the production of blue pigmented actinorhodin (Act) and prodigiosins (e.g. undecylprodigiosin, Red). These genes are divergently encoded and their promoter regions overlap 53bp, as has been shown by gel retardation assays and DNase I footprinting studies. \citep{takano2001complex} Complementary to these findings, the discovery of cis asRNA has also been reported in different studies. \citep{d2010noncoding,panek2008biocomputational,swiercz2008small} The regulatory role of the system's overlapping topology and of the antisense RNA in biological decision making has been demonstrated in number of studies concerning different bacteria, such as ''E. faecalis'' \citep{chatterjee2011convergent} and ''S. enterica''. \citep{lee2010antisense} It is therefore possible that the formation of an antisense RNA between the ''scbA'' and ''scbR'' transcripts could play an important role in the regulation of the GBL system. | ||
| + | |||
| + | Transcription analyses have shown that both genes are mainly active during transition from logarithmic growth to stationary phase. It is presumed that SCBs slowly accumulate into the media and upon reaching a concentration threshold promote a coordinated switch-like transition to antibiotic production by binding to ScbR. However, the mechanism of this network is not fully defined, although several alternative scenarios have been proposed. \citep{Takano2006287,Mehra2008,Chatterjee2011} The complete elucidation of this system could potentially lead to the design of robust and sensitive systems with significant applications as orthologous regulatory circuits in synthetic biology and biotechnology. \cite{BiarnesCarrera201591} | ||
== References == | == References == | ||
<references/> | <references/> | ||
Revision as of 13:46, 3 July 2019
Bacterial communication and Synthetic Biology
Many microorganisms produce antibiotics in order to gain a competitive advantage over other organisms for their survival. However, coordination between members of the population is critical as the end products can be fatal for the colony if produced in an unregulated manner. One prominent mechanism employed is a mechanism of "voting" by members of the colony, by sensing and producing signalling molecules. If a large number of surrounding neigbours are producing the signalling molecules in response to an environmental cue, this is likely to indicate the right moment to produce antibiotics. [1]
These mechanisms have been targeted by synthetic biology whose core aim is the design and engineering of complex biological systems with functionalities that do not exist in nature. In order to accomplish this, novel regulatory circuits need to be developed, which will enable the precise control of gene expression over a wide range of conditions. [2] The bacterium Vibrio fischeri has been in the spotlight in recent years, due to its communication mechanism known as "quorum sensing" (QS). [3] This organism produces acyl homoserine lactone (AHL) as a signal molecule and exports it to the environment. As the colony grows and the cell density increases, the concentration of AHL in the environment also rises, until it reaches a threshold which activates the expression of specific genes responsible for the emission of light.
The system responsible for the production of AHL is composed of two genes and their encoding regulatory proteins LuxI and LuxR. LuxI is the actual synthase of the autoinducer and LuxR, is a repressor belonging to the TetR family. [4] AHL forms a complex with the LuxR protein and together they bind to a short sequence called lux box which both enhances the transcription of LuxI (thus leading to even further accumulation of AHL) and activates the bioluminescence gene cluster. This positive feedback loop leads to a behaviour similar to a bistable switch, with the system being either in the "on" or in the "off" state, and with a zone of unstable intermediate states between them.
Regulation in Streptomyces coelicolor
Although the quorum sensing circuit has been widely employed in synthetic biology with numerous successful applications, \citep{you2004syn,danino2010,sayut2009} it has some important limitations, such as potential crosstalk between different systems due to the promiscuity of the signalling molecules or the promoters, \citep{wu2014quorum} and the problematic implementation in eukaryotic organisms. \citep{hartmann2012} There is therefore a need to expand the available potential circuits which will enhance the synthetic biology toolbox and pave the way to even more exciting applications.
A good candidate for this purpose could be the γ-butyrolactone (GBL) signalling circuits of Streptomyces coelicolor. These are Gram-positive, filamentous, soil-dwelling bacteria, which are known as prolific source of secondary metabolites, such as antibiotics (actinorhodin (Act), undecylprodigiosin (Red), methylenomycin (Mmy), and calcium-dependent antibiotic (CDA)). \citep{Cimermancic2014412,price1999strept} As the end-products can be toxic even to the producing organisms, \citep{Takano2006287} antibiotic production is carefully coordinated in the bacterial population. One way \textit{Streptomyces} can regulate secondary metabolite production is through the use of small diffusible molecules, known as γ-butyrolactones, in a manner analogous to acyl homoserine lactone (AHL)-based quorum sensing (QS). \citep{Ng2009197} At this point, more than fifteen different GBLs have been identified in various Streptomyces species. \citep{Takano2006287,kitani2011avenolide}
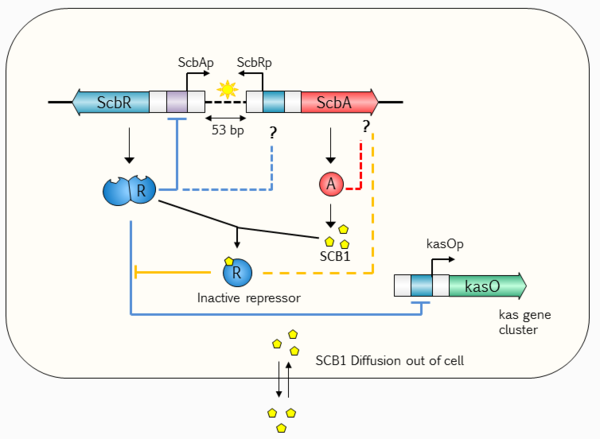
Recent studies and reviews \citep{VanWezel20111311,Liu2013112} point to the hypothesis that the GBL regulatory system involves a small, yet complex two gene network composed by a synthase (scbA (SCO6266)) and a butyrolactone receptor (scbR (SCO6265)), which governs a potentially bistable switch between the "on" and "off" states of antibiotic production. The scbA gene product is presumed to catalyse the condensation of dihydroxyacetone phosphate with a beta--ketoacid to produce three different butyrolactones (SCB1, SCB2 and SCB3). \citep{Takano2006287,kato2007biosynthesis} On the other hand, scbR is a TetR-like \citep{Ramos2005326} DNA-binding protein known to regulate its own transcription and that of scbA. Additionally, ScbR protein has a γ-butyrolactone binding domain at its C-terminal and a DNA binding domain at its N-terminal \citep{natsume2004crystal}. This structure allows it to directly regulate the production of a cryptic metabolite and indirectly the production of blue pigmented actinorhodin (Act) and prodigiosins (e.g. undecylprodigiosin, Red). These genes are divergently encoded and their promoter regions overlap 53bp, as has been shown by gel retardation assays and DNase I footprinting studies. \citep{takano2001complex} Complementary to these findings, the discovery of cis asRNA has also been reported in different studies. \citep{d2010noncoding,panek2008biocomputational,swiercz2008small} The regulatory role of the system's overlapping topology and of the antisense RNA in biological decision making has been demonstrated in number of studies concerning different bacteria, such as E. faecalis \citep{chatterjee2011convergent} and S. enterica. \citep{lee2010antisense} It is therefore possible that the formation of an antisense RNA between the scbA and scbR transcripts could play an important role in the regulation of the GBL system.
Transcription analyses have shown that both genes are mainly active during transition from logarithmic growth to stationary phase. It is presumed that SCBs slowly accumulate into the media and upon reaching a concentration threshold promote a coordinated switch-like transition to antibiotic production by binding to ScbR. However, the mechanism of this network is not fully defined, although several alternative scenarios have been proposed. \citep{Takano2006287,Mehra2008,Chatterjee2011} The complete elucidation of this system could potentially lead to the design of robust and sensitive systems with significant applications as orthologous regulatory circuits in synthetic biology and biotechnology. \cite{BiarnesCarrera201591}
References
- ↑ S. Mehra, S. Charaniya, E. Takano, and W.-S. Hu. A bistable gene switch for antibiotic biosynthesis: The butyrolactone regulon in streptomyces coelicolor. PLoS ONE, 3(7), 2008.
- ↑ M. Biarnes-Carrera, R. Breitling, E. Takano Butyrolactone signalling circuits for synthetic biology. Current Opinion in Chemical Biology 28: 91-98, 2015.
- ↑ Z. Li and S. K. Nair, Quorum sensing: how bacteria can coordinate activity and synchronize their response to external signals? Protein Science 21(10): 1403-1417, 2012.
- ↑ Ramos JL, Martínez-Bueno M, Molina-Henares AJ, et al. , The TetR Family of Transcriptional Repressors Microbiol Mol Biol Rev., 69(2): 326–356, 2005.