Antisense interaction between r and a
The full length mRNA of scbR (r) binds to the full length mRNA of scbA (a) and form a complex which prevents further translation of both mRNAs.
Contents
Chemical equation
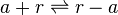
Rate equation
Parameters
The parameters of this reaction is the binding and unbinding rate constant for the interaction of scbR mRNA with scbA mRNA (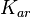 ) and (
) and (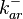 ). The parameter values are derived from various RNA/RNA interaction studies in bacteria.
). The parameter values are derived from various RNA/RNA interaction studies in bacteria.
| Name | Value | Units | Value in previous GBL models [1] | Remarks-Reference |
|---|---|---|---|---|
|
Failed to parse (Cannot store math image on filesystem.): 0.20-89 [2] [3] [4] [5] [6] | 
|
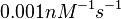 [1] [1]
(Bistability range: |
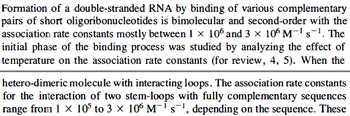
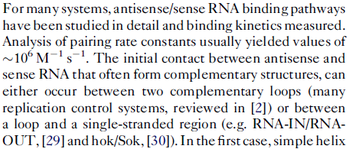
In a 1991 review, Y. Eguchi reported association rates for double stranded RNA binding in the range of 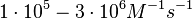 . A similar range (~ . A similar range (~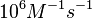 ) is published by S. Brantl in a 2007 review on regulatory mechanisms employed by antisense RNAs. ) is published by S. Brantl in a 2007 review on regulatory mechanisms employed by antisense RNAs.
These values are also in agreement with a study by Franch et al. on the effect of a U-turn loop structure in RNA/RNA interactions, who reported binding rate constants Finally, Nordgren et al. published a study on the kinetics of antisense RNA interactions by using Surface Plasmon Resonance (in wild-type and mutant RNA pairs), where the values reported for the association rate constants are in the range of 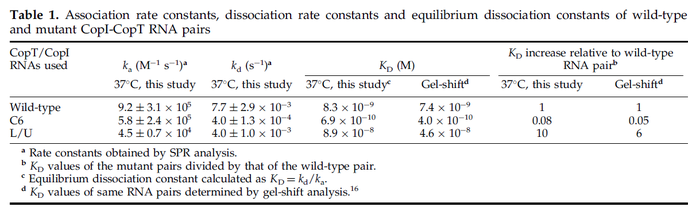 Nordgren et al. 2001[6] Assuming that the dissociation rate is in the range of |
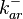
|
Failed to parse (Cannot store math image on filesystem.): 0.006-6 [6] [7] | 
|
Failed to parse (Cannot store math image on filesystem.): 0.01 nM^{-1} s^{-1}
[1]
(Bistability range: |
In the study by Nordgren et al., dissociation rate constants in the range Failed to parse (Cannot store math image on filesystem.): 4 \cdot 10^{-4}-7.7 \cdot 10^{-3} s^{-1} (0.024-0.0462 min^{-1})
were reported (see figure above). These values are also consistent with the ones published by Lima et al. in a study on antisense oligonucleotide hybridization kinetics, who reported dissociation rate constant values in the range 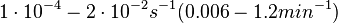
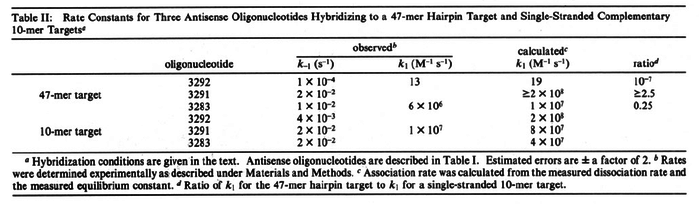 Lima et al. 2001[7] Although the values retrieved by literature are between |
Parameters with uncertainty
When deciding how to describe the uncertainty for these parameter we must take into consideration the fact that many of the reported values are derived from in vitro experiments and correspond to mRNA sequences or fragments of different bacteria species. This means that there might be some difference between actual parameter values and the ones reported in literature. These facts influence the quantification of the parameter uncertainty and therefore the shape of the corresponding distribution.
With regards to the 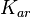 the values that are mostly encountered in different publications are in the range
the values that are mostly encountered in different publications are in the range 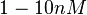 , therefore we put the weight of the distribution in this range and we consider as least likely the larger values as they correspond to mutant RNAs. Therefore, the mode of the log-normal distribution is set to
, therefore we put the weight of the distribution in this range and we consider as least likely the larger values as they correspond to mutant RNAs. Therefore, the mode of the log-normal distribution is set to 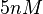 and the confidence interval factor is
and the confidence interval factor is 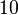 . Thus the range where 95% of the values are found is between Failed to parse (Cannot store math image on filesystem.): 0.5
and Failed to parse (Cannot store math image on filesystem.): 50
.
. Thus the range where 95% of the values are found is between Failed to parse (Cannot store math image on filesystem.): 0.5
and Failed to parse (Cannot store math image on filesystem.): 50
.
Similarly, the values reported for 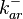 are within the range
are within the range 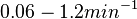 , we put the weight of the distribution within this range but also explore the whole range of values. Therefore, the mode of the log-normal distribution is set to Failed to parse (Cannot store math image on filesystem.): 0.6
and the confidence interval factor is
, we put the weight of the distribution within this range but also explore the whole range of values. Therefore, the mode of the log-normal distribution is set to Failed to parse (Cannot store math image on filesystem.): 0.6
and the confidence interval factor is 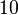 . Thus the range where 95% of the values are found is between
. Thus the range where 95% of the values are found is between 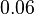 and Failed to parse (Cannot store math image on filesystem.): 6 nM
.
and Failed to parse (Cannot store math image on filesystem.): 6 nM
.
The probability distributions for the two parameters, adjusted accordingly in order to reflect the above values, are the following:
The location and scale parameters of the distribution are:
| Parameter | μ | σ |
|---|---|---|
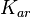
|
Failed to parse (Cannot store math image on filesystem.): 2.4246 | 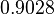
|
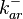
|
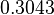
|
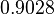
|
References
- ↑ 1.0 1.1 1.2 A. Chatterjee, L. Drews, S. Mehra, E. Takano, Y.N. Kaznessis, and W.-S. Hu. Convergent transcription in the butyrolactone regulon in streptomyces coelicolor confers a bistable genetic switch for antibiotic biosynthesis. PLoS ONE, 6(7), 2011.
- ↑ Shokeen S, Johnson CM, Greenfield TJ, Manias DA, Dunny GM, Weaver KE. Structural analysis of the Anti-Q-Qs interaction: RNA-mediated regulation of E. faecalis plasmid pCF10 conjugation. Plasmid. 2010;64(1):26-35.
- ↑ 3.0 3.1 Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Cell regulation (RNA special issue). 2007; 10: 102–109.
- ↑ 4.0 4.1 Eguchi Y., Itoh T., Tomizawa J. Antisense RNA. Annual Review of Biochemistry. 1991;60: 631–652.
- ↑ 5.0 5.1 5.2 Franch T., Petersen M., Wagner EG., Jacobsen JP., Gerdes K. Antisense RNA regulation in prokaryotes: rapid RNA/RNA interaction facilitated by a general U-turn loop structure. J Mol Biol. 1999 Dec 17;294(5):1115-25.
- ↑ 6.0 6.1 6.2 Nordgren S., Slagter-Jäger J.G., Wagner G.H. Real time kinetic studies of the interaction between folded antisense and target RNAs using surface plasmon resonance. J. Mol. Biol. 2001;310(5):1125-34.
- ↑ 7.0 7.1 Lima W.F., Monia B.P., Ecker D.J., Freier S. M. Implication of RNA structure on antisense oligonucleotide hybridization kinetics. Biochemistry 1992 31 (48), 12055-12061


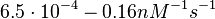 )
)
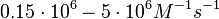 and suggested that ~
and suggested that ~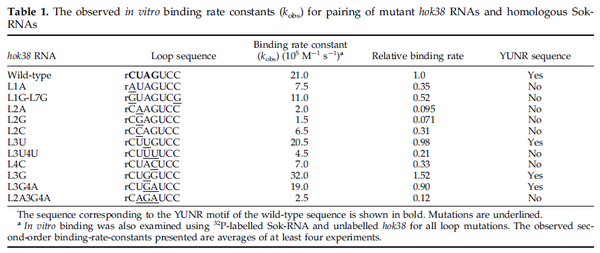

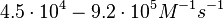 and the resulting equilibrium dissociation constant are in the range of Failed to parse (Cannot store math image on filesystem.): 8.3 \cdot 10^{-8}-6.9 \cdot 10^{-10} M
(Failed to parse (Cannot store math image on filesystem.): 8.3-89 nM
).
and the resulting equilibrium dissociation constant are in the range of Failed to parse (Cannot store math image on filesystem.): 8.3 \cdot 10^{-8}-6.9 \cdot 10^{-10} M
(Failed to parse (Cannot store math image on filesystem.): 8.3-89 nM
).
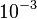 , then from the aforementioned association constants, we calculated the equilibrium dissociation constant as per Failed to parse (Cannot store math image on filesystem.): K_D= /frac{k_d}{k_a}
. So for the range
, then from the aforementioned association constants, we calculated the equilibrium dissociation constant as per Failed to parse (Cannot store math image on filesystem.): K_D= /frac{k_d}{k_a}
. So for the range 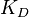 s are Failed to parse (Cannot store math image on filesystem.): 0.2-6.67 nM
.
s are Failed to parse (Cannot store math image on filesystem.): 0.2-6.67 nM
.
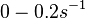 )
)
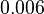 and
and 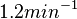 , we wish to explore a wider range of values (
, we wish to explore a wider range of values (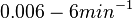 ) in order to take into account the possibility that high GC contents may affect the kinetic rate constants.
) in order to take into account the possibility that high GC contents may affect the kinetic rate constants.