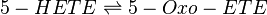Return to overview
The metabolism of 5-HETE to 5-Oxo-ETE is catalysed by the enzyme 5-Hydroxyeicosanoid dehydrogenase (5-HEDH). This enzyme acts reversibly and has been reported to be found in eosinophils, monocytes, DC, B-lymphocytes, keratinocytes and platelets (Steinhilber book) *skin (need to check). The product binds with the OXE receptor (Hosoi2002) and is 30-100 more bioactive than 5-HETE. 5-Oxo-ETE is a chemoattractant for neutrophils and eosinophils.
Doesn't com up in BRENDA or PAXDB - Skeptical
Reaction
Chemical equation
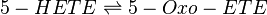
Rate equation
Parameters
Michaelis-Menten Constants
| Value
|
Units
|
Species
|
Notes
|
Reference
|
| 0.0006
|
mM
|
Unkown
|
Follows a ping-pong mechanism by binding with NADP+, transforming it to NADPH and releasing it before binding 5S-HETE.
|
[1]
|
| 0.00067
|
mM
|
Human Cell lines
|
Follows a ping-pong mechanism by binding with NADP+ (Km 139 nM), transforming it to NADPH (inhibited by NADPH (Ki 224 nM)) and releasing it before binding 5S-HETE.
Method: In vitro
Organism: Human
Expression vector: U937 and HL-60 cell lines
Enzyme:5-HEDH
pH: 7.4
Temperature: 37 ◦C
|
[2]
|
| 0.000516 ± 0.00019
|
mM
|
Human Cell lines
|
Method: In vitro
Organism: Human
Expression vector: U937
Enzyme:5-HEDH
pH: 7.4
Temperature: 37 ◦C
|
[3]
|
Michaelis-Menten Constants
| Value
|
Units
|
Species
|
Notes
|
Reference
|
| X
|
Y
|
Z
|
A
|
B
|
Enzyme Turnover Number
| Value
|
Units
|
Species
|
Notes
|
Reference
|
| X
|
Y
|
Z
|
A
|
B
|
5-HEDH Abundance
| Value
|
Units
|
Species
|
Notes
|
Reference
|
| X
|
Y
|
Z
|
A
|
B
|
Gibbs Free energy
| Value
|
Units
|
Species
|
Notes
|
Reference
|
| X
|
Y
|
Z
|
A
|
B
|
Related Reactions