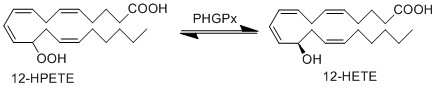
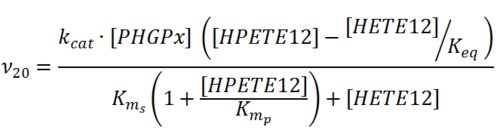Transformation of 12-HPETE to 12-HETE
12-HPETE is reduced by phospholipid hydroperoxide glutathione peroxidase (PHGPx) to the corresponding alcohol, 12-HETE (Suth01). 12-HETE is found in high concentrations in the skin and is renowned for acting as a chemoattractant during inflammation, with binding sites on both LCs and keratinocyte cells (Dowd, Kobza Black et al. 1985).
Contents
Reaction
Chemical equation
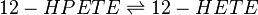
Rate equation
Parameters
Note that the literature values are the same as reaction 12.
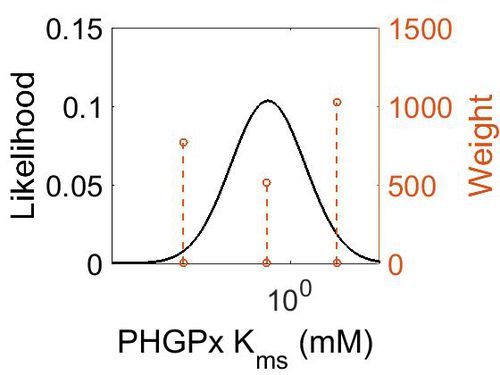
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 3.00E-01 | 
|
Human | Expression Vector: Bioimprited Enzyme - Selenosubtilisin
Enzyme: Glutathione Peroxidase pH: 7 Temperature: 37 |
512 | [1] |
| 11.1 ± 2.90E-01 | 
|
Human | Expression Vector: E Coli
Enzyme: Wild Type Glutathione Peroxidase Enzyme (Se-hGSTZ1-1) pH: 7 Temperature: 37 |
1024 | [2] |
| 4.00E-03 | 
|
Rat | Expression Vector: Rat Liver Cells
Enzyme: Glutathione Peroxidase pH: 7.4 Temperature: 37 |
768 | [3] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 3.19E-01 | 1.02E+03 | 2.54E+00 | 1.92E+00 |
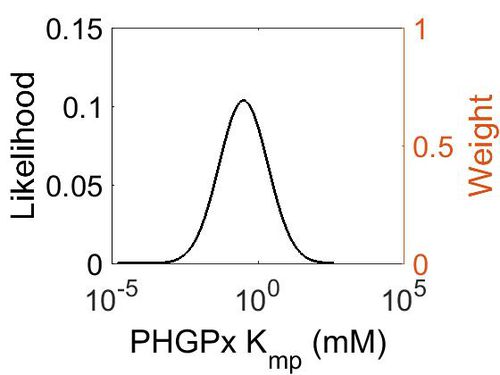
Kmp
This is a “Dependent parameter”, meaning that the log-normal distribution for this parameter was calculated using multivariate distributions (this is discussed in detail here). As a result, no confidence interval factor or literature values were cited for this parameter.
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 3.15E-01 | 2.53E+00 | 1.92E+00 |
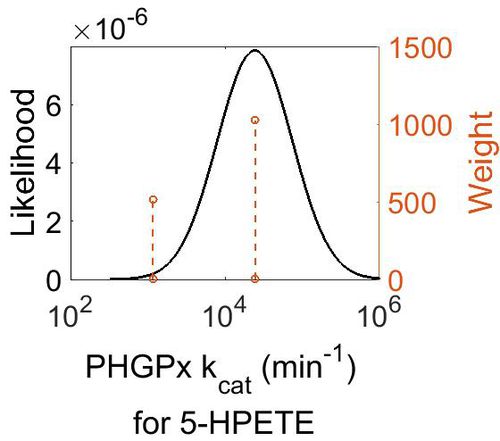
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 1170 ± 50 | 
|
Human | Expression Vector: Bioimprited Enzyme - Selenosubtilisin
Enzyme: Glutathione Peroxidase pH: 7 Temperature: 37 |
512 | [1] |
| 24500 ± 150 | 
|
Human | Expression Vector: E Coli
Enzyme: Wild Type Glutathione Peroxidase Enzyme (Se-hGSTZ1-1) pH: 7 Temperature: 37 |
1024 | [2] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 2.44E+04 | 4.20E+00 | 1.11E+01 | 9.75E-01 |
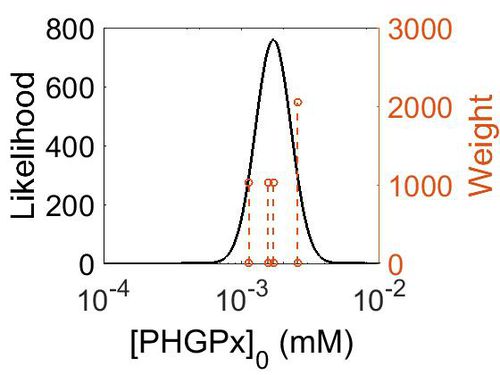
Enzyme concentration
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 459 | 
|
Human | Expression Vector: Skin
Enzyme: PHGPx pH: 7.5 Temperature: 37 °C |
2048 | [4] |
| 307 | 
|
Human | Expression Vector: Lung
Enzyme: PHGPx pH: 7.5 Temperature: 37 °C |
1024 | [4] |
| 282 | 
|
Human | Expression Vector: Esophagus
Enzyme: PHGPx pH: 7.5 Temperature: 37 °C |
1024 | [4] |
| 204 | 
|
Human | Expression Vector: Gut
Enzyme: PHGPx pH: 7.5 Temperature: 37 °C |
1024 | [5] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 3.14E+02 | 1.38E+00 | 5.85E+00 | 3.09E-01 |
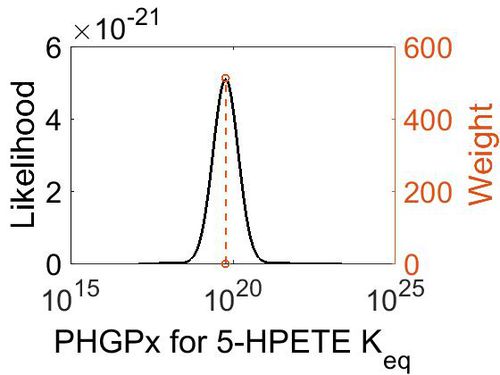
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| (-26.941177) | kcal/mol | Not stated | Estimated
Enzyme: PHGPx Substrate:a hydroperoxy-fatty-acyl-[lipid] Product: a hydroxy-fatty-acyl-[lipid] pH: 7.3 ionic strength: 0.25 |
64 | [6] |
| Mode | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 5.90E+19 | 1.00E+01 | 4.63E+01 | 8.90E-01 |
Related Reactions
- ↑ 1.0 1.1 Liu L. "Functional mimicry of the active site of glutathione peroxidase by glutathione imprinted selenium-containing protein. Biomacromolecules. 2008 Jan;9(1):363-8. doi: 10.1021/bm7008312. Epub 2007 Dec 29.
- ↑ 2.0 2.1 Zheng K. "A novel selenium-containing glutathione transferase zeta1-1, the activity of which surpasses the level of some native glutathione peroxidases. Int J Biochem Cell Biol. 2008;40(10):2090-7. doi: 10.1016/j.biocel.2008.02.006. Epub 2008 Feb 15.
- ↑ Hiratsuka A. "Subunit Ya-specific glutathione peroxidase activity toward cholesterol 7-hydroperoxides of glutathione S-transferases in cytosols from rat liver and skin. J Biol Chem. 1997 Feb 21;272(8):4763-9.
- ↑ 4.0 4.1 4.2 M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587
- ↑ M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471