Difference between revisions of "Kinetic model of Central Metabolism"
| Line 23: | Line 23: | ||
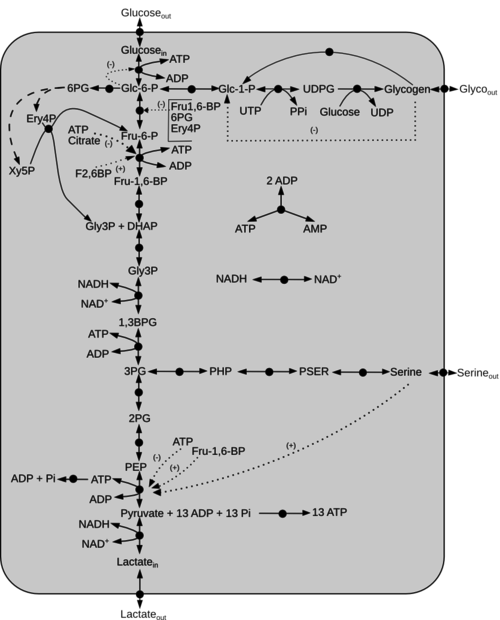
rect 320 670 350 700 [[PDH|Phosphoglycerate dehydrogenase]] | rect 320 670 350 700 [[PDH|Phosphoglycerate dehydrogenase]] | ||
rect 490 670 520 700 [[PSA|Phosphoserine amino-transferase]] | rect 490 670 520 700 [[PSA|Phosphoserine amino-transferase]] | ||
| − | rect | + | rect 670 670 700 700 [[PSP|Phosphohydroxypyruvate]] |
rect 250 705 280 735 [[PGAM|3-phosphoglycerate mutase]] | rect 250 705 280 735 [[PGAM|3-phosphoglycerate mutase]] | ||
rect 250 800 280 830 [[ENO|Enolase]] | rect 250 800 280 830 [[ENO|Enolase]] | ||