Difference between revisions of "Transformation of PGH2 to PGE2"
(→PGES Enzyme Parameters) |
(→PGES Enzyme Parameters) |
||
| Line 80: | Line 80: | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
| − | |+ style="text-align: left;" | | + | |+ style="text-align: left;" | PGES Abundance |
|- | |- | ||
! Value | ! Value | ||
| Line 88: | Line 88: | ||
! Reference | ! Reference | ||
|- | |- | ||
| − | | | + | |220 |
| − | | | + | |<math> ppm </math> |
|Human | |Human | ||
| − | | | + | |Expression Vector: Placenta |
| − | + | Enzyme: PGES | |
| − | |<ref name=" | + | pH: 7.5 |
| + | Temperature: 37 °C | ||
| + | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | ||
| + | human proteome'' Nature, 2014 509, 582–587]</ref> | ||
|- | |- | ||
| − | | | + | |75.3 |
| − | | | + | |<math> ppm </math> |
| − | |Human | + | |Human |
| − | | | + | |Expression Vector: Urinary Bladder |
| − | |<ref name=" | + | Enzyme: PGES |
| + | pH: 7.5 | ||
| + | Temperature: 37 °C | ||
| + | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
| + | |- | ||
| + | |208 | ||
| + | |<math> ppm </math> | ||
| + | |Human | ||
| + | |Expression Vector: Stomach | ||
| + | Enzyme: PGES | ||
| + | pH: 7.5 | ||
| + | Temperature: 37 °C | ||
| + | |<ref name="Wilhelm2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm ''Mass-spectrometry-based draft of the | ||
| + | human proteome'' Nature, 2014 509, 582–587]</ref> | ||
| + | |- | ||
| + | |28.1 | ||
| + | |<math> ppm </math> | ||
| + | |Human | ||
| + | |Expression Vector: Lung | ||
| + | Enzyme: PGES | ||
| + | pH: 7.5 | ||
| + | Temperature: 37 °C | ||
| + | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
| + | |- | ||
| + | |11.6 | ||
| + | |<math> ppm </math> | ||
| + | |Human | ||
| + | |Expression Vector: Colon | ||
| + | Enzyme: PGES | ||
| + | pH: 7.5 | ||
| + | Temperature: 37 °C | ||
| + | |<ref name="Kim2014"> [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13302.pdf M. Kim ''A draft map of the human proteome'' Nature, 2014 509, 575–581]</ref> | ||
|- | |- | ||
|} | |} | ||
Revision as of 10:31, 9 August 2016
Contents
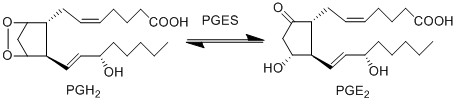
Reaction
Chemical equation
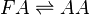
Rate equation
PGES Enzyme Parameters
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 1.6E-01 ± 4.00E-03 | 
|
Human | Expression Vector: E. Coli
Enzyme: PGES pH: 8 Temperature: 37 |
[1] |
| 2.15E-01 | 
|
Human | Wild Type Enzyme | [2] |
| 1.49E-02 | 
|
Human | Expression Vector: E. Coli
Enzyme: PGES pH: Unknown Temperature: Unknown Other: cPGES, casein kinase II and Hsp90 |
[3] |
| 6.66E-02 | 
|
Human | Expression Vector: E. Coli
Enzyme: PGES pH: Unknown Temperature: Unknown |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 3000 ± 360 | per minute | Human | Expression Vector: E. Coli.
Enzyme: Microsomal Prostaglandin E Synthase pH: 7.5 Temperature: 37 |
[4] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 220 | 
|
Human | Expression Vector: Placenta
Enzyme: PGES pH: 7.5 Temperature: 37 °C |
[5] |
| 75.3 | 
|
Human | Expression Vector: Urinary Bladder
Enzyme: PGES pH: 7.5 Temperature: 37 °C |
[6] |
| 208 | 
|
Human | Expression Vector: Stomach
Enzyme: PGES pH: 7.5 Temperature: 37 °C |
[5] |
| 28.1 | 
|
Human | Expression Vector: Lung
Enzyme: PGES pH: 7.5 Temperature: 37 °C |
[6] |
| 11.6 | 
|
Human | Expression Vector: Colon
Enzyme: PGES pH: 7.5 Temperature: 37 °C |
[6] |
References
- ↑ Pettersson P. , "Identification of beta-trace as prostaglandin D synthase. FASEB J. 2010 Dec;24(12):4668-77. doi: 10.1096/fj.10-164863. Epub 2010 Jul 28.
- ↑ Hamza A. , "Understanding microscopic binding of human microsomal prostaglandin E synthase-1 (mPGES-1) trimer with substrate PGH2 and cofactor GSH: insights from computational alanine scanning and site-directed mutagenesis. J Phys Chem B. 2010 Apr 29;114(16):5605-16. doi: 10.1021/jp100668y.
- ↑ 3.0 3.1 Kobayashi T. , "Regulation of cytosolic prostaglandin E synthase by phosphorylation. Biochem J. 2004 Jul 1;381(Pt 1):59-69.
- ↑ [www.ncbi.nlm.nih.gov/pubmed/16399384 Pettersson P., "Human microsomal prostaglandin E synthase 1: a member of the MAPEG protein superfamily. Methods Enzymol. 2005;401:147-61.]
- ↑ 5.0 5.1 [http://www.nature.com/nature/journal/v509/n7502/pdf/nature13319.pdf M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587]
- ↑ 6.0 6.1 6.2 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581