Difference between revisions of "Phosphofructokinase type 1"
(→Parameters with uncertainty) |
(→Parameters with uncertainty) |
||
| Line 83: | Line 83: | ||
*<math>Km_{ADP} = 1.4</math> for ''Thermotoga maritima'' is being reported in Hansen, T., M. Musfeldt ''et. al.'' <ref name="Hansen">Hansen, T., M. Musfeldt, and P. Schonheit (2002), ''ATP-dependent 6-phosphofructokinase from the hyperthermophilic bacterium Thermotoga maritima: characterization of an extremely thermophilic, allosterically regulated enzyme''. Arch. Microbiol. 177:401-409 </ref> and <math>Km_{Fru1,6BP} = 16.7</math> for ''Desulfurococcus amylolyticus'' is reported in Hansen T, Schönheit P. ''et. al.''<ref name=Hansen_2003">Hansen T, Schönheit P. (2003),''Purification and Characterization of the MQH2:NO Oxidoreductase from the Hyperthermophilic Archaeon Pyrobaculum aerophilum'', J Biol Chem, 278 (38), 35861-35868 </ref>. The mean and std. dev. is calculated as <math>0.945 \pm 0.454</math> | *<math>Km_{ADP} = 1.4</math> for ''Thermotoga maritima'' is being reported in Hansen, T., M. Musfeldt ''et. al.'' <ref name="Hansen">Hansen, T., M. Musfeldt, and P. Schonheit (2002), ''ATP-dependent 6-phosphofructokinase from the hyperthermophilic bacterium Thermotoga maritima: characterization of an extremely thermophilic, allosterically regulated enzyme''. Arch. Microbiol. 177:401-409 </ref> and <math>Km_{Fru1,6BP} = 16.7</math> for ''Desulfurococcus amylolyticus'' is reported in Hansen T, Schönheit P. ''et. al.''<ref name=Hansen_2003">Hansen T, Schönheit P. (2003),''Purification and Characterization of the MQH2:NO Oxidoreductase from the Hyperthermophilic Archaeon Pyrobaculum aerophilum'', J Biol Chem, 278 (38), 35861-35868 </ref>. The mean and std. dev. is calculated as <math>0.945 \pm 0.454</math> | ||
| − | * Similarly for <math>Km_{Fru1,6BP} = 7.6</math> | + | * Similarly for <math>Km_{Fru1,6BP} = 7.6</math> in ''Thermotoga maritima'' is being reported in Hansen, T., M. Musfeldt ''et. al.'' <ref name="Hansen"></ref> and <math>Km_{ADP} = 0.49</math> in ''Desulfurococcus amylolyticus'' is reported in Hansen T, Schönheit P. ''et. al.''<ref name="Hansen_2003"></ref>. The mean and std. dev. is calculated as <math>12.15 \pm 4.54</math> |
* Four Keq values have been reported in the SilicoTrypWiki (Wikipedia for insilico modelling of Trypanosome) for Phosphofructokinase: 308.4, 254, 1035, 800. Taking the mean and std. dev. of this value is <math> 599.35 \pm 329.38</math> <ref name="SilicoTryp">[[http://silicotryp.ibls.gla.ac.uk/wiki/Phosphofructokinase Silicotryp]]</ref> | * Four Keq values have been reported in the SilicoTrypWiki (Wikipedia for insilico modelling of Trypanosome) for Phosphofructokinase: 308.4, 254, 1035, 800. Taking the mean and std. dev. of this value is <math> 599.35 \pm 329.38</math> <ref name="SilicoTryp">[[http://silicotryp.ibls.gla.ac.uk/wiki/Phosphofructokinase Silicotryp]]</ref> | ||
Revision as of 11:20, 25 April 2014
The enzyme Phosphofructokinase Type-1 uses another ATP molecule to transfer a phosphate group to Fru6P to form fructose 1, 6-bisphosphate. PFK-1 is an allosteric enzyme showing cooperative behaviour with Fru6P and hyperbolic kinetics with ATP.
Contents
Chemical equation
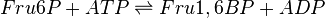
Rate equation
The concerted transition model of Monod, Wyman and Changeux (MWC model) is used as a rate equation for this tetrameric enzyme for considering exclusive ligand binding (F6P, activators and inhibitors) together with mixed type activation, (Fru2,6BP or AMP or Pi) [1].
Parameter values
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
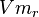
|
0.031 [1] | 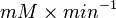
|
HeLa cell line | Moreno-Sánchez, Marín-Hernández, Encalada & Saavedra, unpublished results |
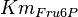
|
1.0 [1] | mM | ||
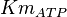
|
0.021[1] | mM | ||
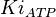
|
20[1] | mM | ||
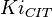
|
6.8[1] | mM | ||
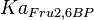
|
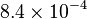 [1] [1]
|
mM | ||
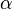
|
0.32[1] | Dimensionless | ||
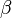
|
0.98[1] | Dimensionless | ||
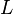
|
4.1[1] | Dimensionless | ||
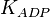
|
5[1] | mM | ||
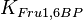
|
5[1] | mM | ||

|
247[1] | mM | Recalculated from the 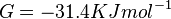
|
Parameters with uncertainty
- The
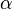 and
and 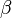 in the rate equation represents the factors by which the ligand affinity and catalytic capacity are modified in the presence of an allosteric activatory [2]. As
in the rate equation represents the factors by which the ligand affinity and catalytic capacity are modified in the presence of an allosteric activatory [2]. As 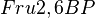 is the only activator in our model we considered the
is the only activator in our model we considered the 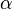 and
and 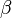 value of
value of 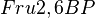 ,
, 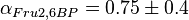 and
and 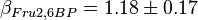 . These two values are measured in the presence of 140
. These two values are measured in the presence of 140 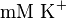 medium [2].
medium [2].
- The Vm value is reported as Failed to parse (Cannot store math image on filesystem.): 56 \pm 23
(nmol/min/mg protein). HeLa cells were harvested at a concentration of 65 mg protein/ml [2]. Converting Vm to mM/min becomes Failed to parse (Cannot store math image on filesystem.): 0.00364 \pm 0.001495
again in the presence of 140
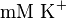 .
.
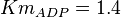 for Thermotoga maritima is being reported in Hansen, T., M. Musfeldt et. al. [3] and
for Thermotoga maritima is being reported in Hansen, T., M. Musfeldt et. al. [3] and 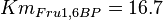 for Desulfurococcus amylolyticus is reported in Hansen T, Schönheit P. et. al.[4]. The mean and std. dev. is calculated as
for Desulfurococcus amylolyticus is reported in Hansen T, Schönheit P. et. al.[4]. The mean and std. dev. is calculated as 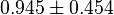
- Similarly for
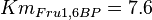 in Thermotoga maritima is being reported in Hansen, T., M. Musfeldt et. al. [3] and
in Thermotoga maritima is being reported in Hansen, T., M. Musfeldt et. al. [3] and 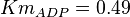 in Desulfurococcus amylolyticus is reported in Hansen T, Schönheit P. et. al.[5]. The mean and std. dev. is calculated as
in Desulfurococcus amylolyticus is reported in Hansen T, Schönheit P. et. al.[5]. The mean and std. dev. is calculated as 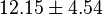
- Four Keq values have been reported in the SilicoTrypWiki (Wikipedia for insilico modelling of Trypanosome) for Phosphofructokinase: 308.4, 254, 1035, 800. Taking the mean and std. dev. of this value is
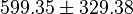 [6]
[6]
| Parameter | Value | Units |
|---|---|---|
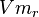
|
Failed to parse (Cannot store math image on filesystem.): 0.00364 \pm 0.001495 | 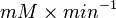
|
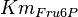
|
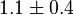
|
mM |
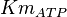
|
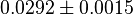 [2] [2]
|
mM |
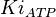
|
20[1] | mM |
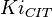
|
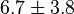
|
mM |
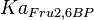
|
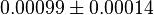
|
mM |
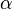
|
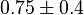
|
Dimensionless |
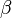
|
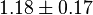
|
Dimensionless |
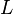
|
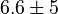
|
Dimensionless |
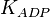
|
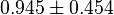
|
mM |
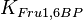
|
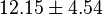
|
mM |

|
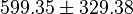
|
References
- ↑ 1.00 1.01 1.02 1.03 1.04 1.05 1.06 1.07 1.08 1.09 1.10 1.11 1.12 1.13 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011). Modeling cancer glycolysis. Biochim Biophys Acta, 1807:755–767 (doi)
- ↑ 2.0 2.1 2.2 2.3 R. Moreno-Sánchez, A. Marín-Hernández, J.C. Gallardo-Pérez, H. Quezada, R. Encalada, S. Rodríguez-Enríquez et al. (2012), Phosphofructokinase type 1 kinetics, isoform expression, and gene polymorphisms in cancer cells, J Cell Biochem, 113, pp. 1692–1703
- ↑ 3.0 3.1 Hansen, T., M. Musfeldt, and P. Schonheit (2002), ATP-dependent 6-phosphofructokinase from the hyperthermophilic bacterium Thermotoga maritima: characterization of an extremely thermophilic, allosterically regulated enzyme. Arch. Microbiol. 177:401-409
- ↑ Hansen T, Schönheit P. (2003),Purification and Characterization of the MQH2:NO Oxidoreductase from the Hyperthermophilic Archaeon Pyrobaculum aerophilum, J Biol Chem, 278 (38), 35861-35868
- ↑ Cite error: Invalid
<ref>tag; no text was provided for refs namedHansen_2003 - ↑ [Silicotryp]