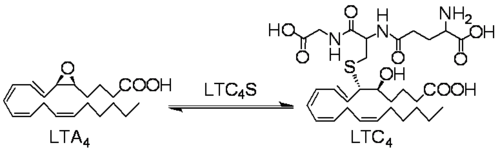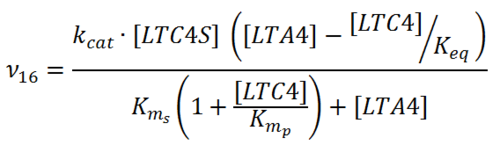Difference between revisions of "Transformation of LTA4 to LTC4"
(→Parameters) |
|||
| Line 27: | Line 27: | ||
! Reference | ! Reference | ||
|- | |- | ||
| − | |0.3 | + | |0.3 ± 0.06 |
|mM | |mM | ||
|Human | |Human | ||
| Line 46: | Line 46: | ||
Temperature: 37 °C | Temperature: 37 °C | ||
|1024 | |1024 | ||
| − | |<ref name="Niegowski2013"> [http://www.jbc.org/content/early/2013/12/23/jbc.M113.534628 Niegowski D. " Crystal structures of Leukotriene C4 synthase in complex with product analogs, implications for the | + | |<ref name="Niegowski2013"> [http://www.jbc.org/content/early/2013/12/23/jbc.M113.534628 Niegowski D. " Crystal structures of Leukotriene C4 synthase in complex with product analogs, implications for the enzyme mechanism'' J. Biol. Chem. 289, 5199-5207 (2014)]</ref> |
| − | enzyme mechanism'' J. Biol. Chem. 289, 5199-5207 (2014)]</ref> | ||
|- | |- | ||
|} | |} | ||
| Line 100: | Line 99: | ||
Temperature: 37 °C | Temperature: 37 °C | ||
|1024 | |1024 | ||
| − | |<ref name="Niegowski2013"> [http://www.jbc.org/content/ | + | |<ref name="Niegowski2013"> [http://www.jbc.org/content/early/2013/12/23/jbc.M113.534628 Niegowski D. " Crystal structures of Leukotriene C4 synthase in complex with product analogs, implications for the enzyme mechanism'' J. Biol. Chem. 289, 5199-5207 (2014)]</ref> |
| − | enzyme mechanism'' J. Biol. Chem. 289, 5199-5207 (2014)]</ref> | ||
|- | |- | ||
|} | |} | ||
Latest revision as of 10:12, 2 November 2019
Leukotriene C4 synthase (LTC4S) transforms LTA4 into LTC4. This reaction involves the conjugation of the LTA4 epoxide and glutathione.
Contents
Reaction
Chemical equation
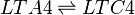
Rate equation
Parameters
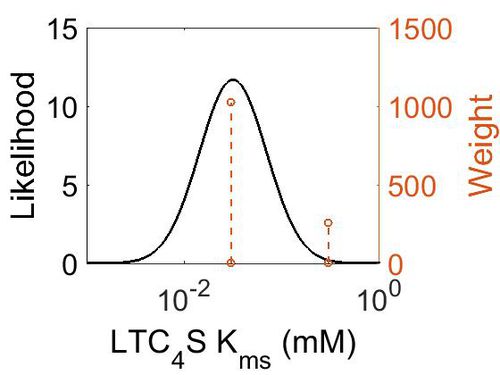
Kms
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 0.3 ± 0.06 | mM | Human | Expression Vector: E. Coli.
Enzyme: Wild Type hLTC4S pH:7.8 Temperature:20 °C |
256 | [1] |
| 3.00E-02 ± 1.00E-02 | mM | Human | Expression Vector: E Coli
Enzyme: Wild type LTC4S pH: 7.8 Temperature: 37 °C |
1024 | [2] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 3.15E-02 | 7.18E+00 | -2.83E+00 | 7.90E-01 |
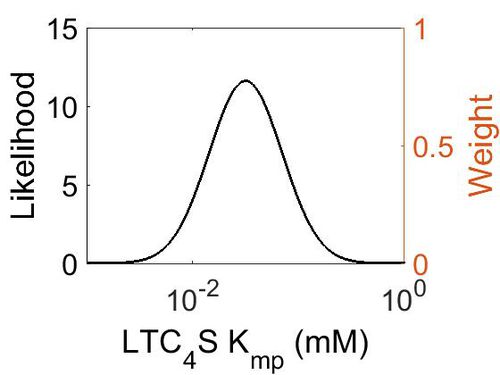
Kmp
| Mode (mM) | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|
| 3.18E-02 | -2.82E+00 | 7.92E-01 |
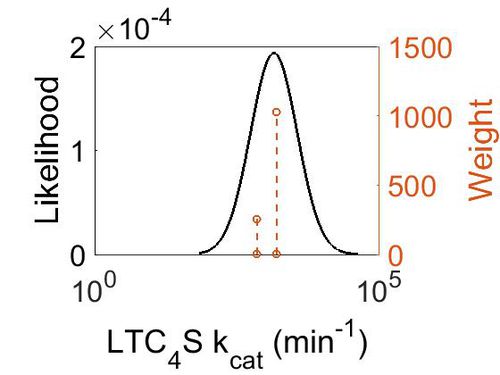
kcat
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 702 | per minute | Human | Expression Vector: E. Coli.
Enzyme: Wild Type hLTC4S pH:7.8 Temperature:20 °C |
256 | [1] |
| 1560 ± 240 | per minute | Human | Expression Vector: E Coli
Enzyme: Wild type LTC4S pH: 7.8 Temperature: 37 °C |
1024 | [2] |
| Mode (min-1) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 1.47E+03 | 1.42E+00 | 7.40E+00 | 3.29E-01 |
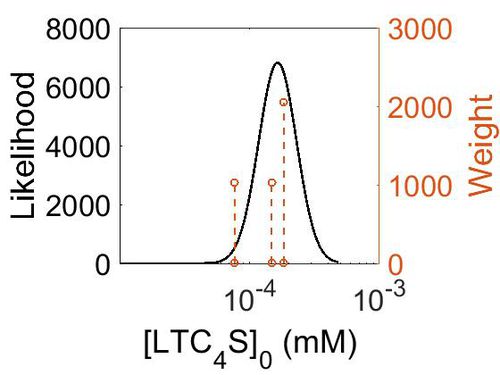
Enzyme concentration
To convert the enzyme concentration from ppm to mM, the following equation was used.
| Value | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 26.8 | 
|
Human | Expression Vector: Lung
Enzyme: LTC4S pH: 7.5 Temperature: 37 °C |
1024 | [3] |
| 33.0 | 
|
Human | Expression Vector: Esophagus
Enzyme: LTC4S pH: 7.5 Temperature: 37 °C |
2048 | [3] |
| 13.9 | 
|
Human | Expression Vector: Adrenal Gland
Enzyme: LTC4S pH: 7.5 Temperature: 37 °C |
1024 | [3] |
| Mode (mM) | Mode (ppm) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|---|
| 2.90E+01 | 1.60E-04 | 1.44E+00 | 3.49E+00 | 3.46E-01 |
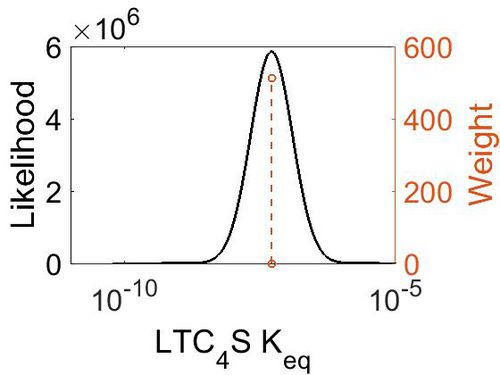
Keq
| Gibbs Free Energy Change | Units | Species | Notes | Weight | Reference |
|---|---|---|---|---|---|
| 9.934128 | kcal/mol | Not stated | Estimated
Enzyme: LTC4S Substrate: LTA4 Product: LTC4 pH: 7.3 ionic strength: 0.25 |
64 | [4] |
| Mode (mM) | Confidence Interval | Location parameter (µ) | Scale parameter (σ) |
|---|---|---|---|
| 5.13E-08 | 1.00E+01 | -1.60E+01 | 8.90E-01 |
Related Reactions
References
- ↑ 1.0 1.1 [http://www.jbc.org/content/285/52/40771.full.pdf Rinaldo A. " Arginine 104 Is a Key Catalytic Residue in Leukotriene C4 Synthase J Biochem 2010, 285, 40771-40776]
- ↑ 2.0 2.1 Niegowski D. " Crystal structures of Leukotriene C4 synthase in complex with product analogs, implications for the enzyme mechanism J. Biol. Chem. 289, 5199-5207 (2014)
- ↑ 3.0 3.1 3.2 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ Caspi et al 2014, "The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of Pathway/Genome Databases," Nucleic Acids Research 42:D459-D471