Difference between revisions of "UDPG-pyrophosphorylase"
(→Equilibrium constant) |
|||
| Line 88: | Line 88: | ||
! Source | ! Source | ||
|- | |- | ||
| − | | 0. | + | | <math>0.20 \pm 0.08</math> |
| − | | pH=7, T=25°C, 10mM Mg2+ | + | | pH=7 and 7.9, T=25°C, 10mM Mg2+ |
| − | | NIST database "Thermodynamics of Enzyme-Catalyzed Reactions" entry [[http://xpdb.nist.gov/enzyme_thermodynamics/enzyme_data1.pl?T1=58TUR/TUR_684]] from Atkinson et al. (1958) <ref name="Turner">Turner, D.H.; Turner, J.F.; Biochem. J.; 69, 448 (1958)</ref> | + | | NIST database "Thermodynamics of Enzyme-Catalyzed Reactions" entry [[http://xpdb.nist.gov/enzyme_thermodynamics/enzyme_data1.pl?T1=58TUR/TUR_684]] from Atkinson et al. (1958) <ref name="Turner">Turner, D.H.; Turner, J.F.; Biochem. J.; 69, 448 (1958)</ref> reported 4 values for Keq; 0.119, 0.286, 0.139, 0.263. Taking mean and std. for these values give <math>K_{eq} = 0.20 \pm 0.08 </math>(n=4). |
| − | |||
|} | |} | ||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 11:43, 1 July 2014
This enzyme converts UTP and G1P to UDP-glucose (UDPG) and pyrophosphate (PPi)
Contents
Chemical equation
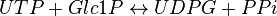
Rate equation
Reversible Bi substrate Michaelis-Menten equation with random binding order is used [1]
![\frac{ \frac{V_{max}}{K_{UTP}K_{Glc1P}} \left( [UTP][Glc1P] - \frac{[UDPG][PPi]}{K_{eq}} \right) }{ \left( 1 + \frac{[UTP]}{K_{UTP}} + \frac{[PPi]}{K_{PPi}} \right) \left( 1 + \frac{[UDPG]}{K_{UDPG}} + \frac{[Glc1P]}{K_{Glc1P}} \right) }](/wiki/images/math/5/c/b/5cb04d31df5c22ab51ad23a54e27d997.png)
Parameter values
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
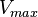
|
200 [2] | 
|
Recombinant, human muscle | |
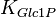
|
0.4 [3] | mM | ||
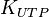
|
0.92 [3] | mM | ||
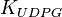
|
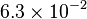 [3] [3]
|
mM | ||
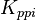
|
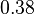 [3] [3]
|
mM | ||

|
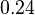 [4] [4]
|
Dimensionless |
Parameters with uncertainty
- The value of
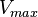 is reported to be
is reported to be 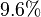 of
of 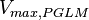 . The Std. Dev. for
. The Std. Dev. for 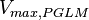 was considered to be
was considered to be 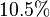 of its mean value. Same error percentage is considered for
of its mean value. Same error percentage is considered for 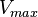 .
.
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
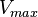
|
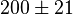 [2] [2]
|

|
Recombinant, human muscle | |
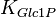
|
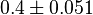 [3] [3]
|
mM | ||
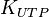
|
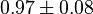 [3] [3]
|
mM | ||
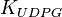
|
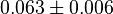 [3] [3]
|
mM | ||
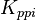
|
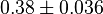 [3] [3]
|
mM | ||

|
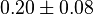
|
Dimensionless |
Equilibrium constant
| Equilibrium constant | Conditions | Source |
|---|---|---|
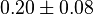
|
pH=7 and 7.9, T=25°C, 10mM Mg2+ | NIST database "Thermodynamics of Enzyme-Catalyzed Reactions" entry [[1]] from Atkinson et al. (1958) [5] reported 4 values for Keq; 0.119, 0.286, 0.139, 0.263. Taking mean and std. for these values give 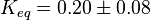 (n=4). (n=4).
|
References
- ↑ Palm, D.C. (2013). The regulatory design of glycogen metabolism in mammalian skeletal muscle (Ph.D.). University of Stellenbosch
- ↑ 2.0 2.1 Villar-Palasi C & Larner J (1960). Levels of activity of the enzymes of the glycogen cycle in rat tissues. Arch Biochem Biophys 86, 270–273.
- ↑ 3.0 3.1 3.2 3.3 3.4 3.5 3.6 3.7 Duggleby RG, Chao YC, Huang JG, Peng HL & Chang HY (1996). Sequence differences between human muscle and liver cDNAs for UDPglucose pyrophosphorylase and kinetic properties of the recombinant enzymes expressed in Escherichia coli. Eur J Biochem 235, 173–179.
- ↑ Bergamini C, Signorini M, Ferrari C & Dallocchio F (1983), Non-Michaelian kinetics of rabbit muscle uridine diphosphoglucose pyrophosphorylase, Arch Biochem Biophys 227, 397–405
- ↑ Turner, D.H.; Turner, J.F.; Biochem. J.; 69, 448 (1958)