Difference between revisions of "Transformation of PGE2 to 15-Keto-PGE2"
(→15-PGDH Parameters) |
(→15-PGDH Parameters) |
||
| Line 83: | Line 83: | ||
Substrate: PGE2 | Substrate: PGE2 | ||
|<ref name="Zhou2001"> [http://onlinelibrary.wiley.com/doi/10.1046/j.1432-1327.2001.02218.x/epdf Zhou H., C-Terminal region of human NAD+-dependent 15-hydroxyprostaglandin dehydrogenase is involved in the interaction with prostaglandin substrates.'' Eur J Biochem. 2001 Jun;268(12):3368-74.]</ref> | |<ref name="Zhou2001"> [http://onlinelibrary.wiley.com/doi/10.1046/j.1432-1327.2001.02218.x/epdf Zhou H., C-Terminal region of human NAD+-dependent 15-hydroxyprostaglandin dehydrogenase is involved in the interaction with prostaglandin substrates.'' Eur J Biochem. 2001 Jun;268(12):3368-74.]</ref> | ||
| + | |- | ||
| + | |0.0055 ± 0.0006 | ||
| + | |<math> mM </math> | ||
| + | |Human | ||
| + | |Method:In vitro | ||
| + | Expression Vector: E. Coli | ||
| + | pH:8 | ||
| + | Temperature: 25 | ||
| + | Substrate: PGE2 + NAD+ | ||
| + | |<ref name="Niesen2010"> [http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0013719 F. Niesen,, High-Affinity Inhibitors of Human NAD+-Dependent 15-Hydroxyprostaglandin Dehydrogenase: Mechanisms of Inhibition and Structure-Activity Relationships'' PLoS One. 2010 Nov 2;5(11):e13719.]</ref> | ||
|- | |- | ||
|} | |} | ||
| Line 96: | Line 106: | ||
! Reference | ! Reference | ||
|- | |- | ||
| − | | | + | |840 |
|<math> per minute </math> | |<math> per minute </math> | ||
| − | | | + | |Human |
| − | | | + | |Method: In vitro |
| − | + | Expression Vector: E. Coli | |
| − | |- | + | pH:8 |
| + | Temperature: 25 | ||
| + | Substrate: PGE2 + NAD+ | ||
| + | |<ref name="Niesen2010"> [http://journals.plos.org/plosone/article?id=10.1371/journal.pone.0013719 F. Niesen,, High-Affinity Inhibitors of Human NAD+-Dependent 15-Hydroxyprostaglandin Dehydrogenase: Mechanisms of Inhibition and Structure-Activity Relationships'' PLoS One. 2010 Nov 2;5(11):e13719.]</ref> | ||
|} | |} | ||
Revision as of 16:23, 28 September 2016
15-hydroxyprostaglandin dehydrogenase, also known as (15-PGDH) metabolises PG into the 15-Keto variant via a ketone reduction. The enzyme is constituently expressed in the skin (Finhelm1982) and within the eicosanoid network it is reported as metabolising PGE2 and PGF2a.
It should be noted that Judson et al found that 15-PGDH expression of the enzyme decreases in response to UVR (Judson2010).
Reaction
Catalysed by 15-PGDH
Chemical equation
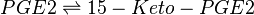
Rate equation
15-PGDH Parameters
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.008 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGE2 + NAD+ |
[1] |
| 0.0075 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGE2 + NADP+ |
[1] |
| 0.024 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGF2a + NAD+ |
[1] |
| 0.023 | 
|
Rat Skin | Method:
pH:7.4 Temperature: 37'C Substrate: PGF2a + NADP+ |
[1] |
| 0.0039 | 
|
Purified Human 15-PGDH | Method: In vitro
Expression Vector: E. coli pH:7.5 Temperature: 37'C Substrate: PGE2 |
[2] |
| 0.0099 | 
|
Purified Rat 15-PGDH | Method: In vitro
Expression Vector: E. coli pH:7.5 Temperature: 37'C Substrate: PGE2 |
[2] |
| 0.0055 ± 0.0006 | 
|
Human | Method:In vitro
Expression Vector: E. Coli pH:8 Temperature: 25 Substrate: PGE2 + NAD+ |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 840 | 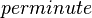
|
Human | Method: In vitro
Expression Vector: E. Coli pH:8 Temperature: 25 Substrate: PGE2 + NAD+ |
[3] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|

|
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
References
- ↑ 1.0 1.1 1.2 1.3 N. Fincham, Novel prostaglandin dehydrogenase in rat skin. Biochem J. 1983 Apr 15;212(1):129-34.
- ↑ 2.0 2.1 Zhou H., C-Terminal region of human NAD+-dependent 15-hydroxyprostaglandin dehydrogenase is involved in the interaction with prostaglandin substrates. Eur J Biochem. 2001 Jun;268(12):3368-74.
- ↑ 3.0 3.1 F. Niesen,, High-Affinity Inhibitors of Human NAD+-Dependent 15-Hydroxyprostaglandin Dehydrogenase: Mechanisms of Inhibition and Structure-Activity Relationships PLoS One. 2010 Nov 2;5(11):e13719.