Serine out
This reaction describes the utilization of the endproduct Serine in other pathways.
Contents
Reaction equation
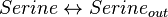
Rate equation
Simple reversible mass action rate law is used.
![v = K_{1} * [Serine] - K_{2} * [Serine_{out}]](/wiki/images/math/e/6/b/e6b7d292763cb679cbd1de648b4b0ddf.png)
Parameters
- For Transport reactions it is presumed to be at equilibrium, such that
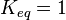 and
and 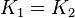 .
.
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
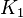
|
0.0103 [1] | 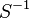
|
Escherichia coli | |
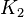
|
0.0103 | 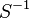
|
Parameters with uncertainty
- The transport rates have been modelled using mass action kinetics (i.e., as non-saturable, non-enzymatic reactions). No information is available about the uncertainty of these parameters. As these parameters are strictly positive, they are sampled using a log-normal distribution as are and values. The means of
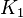 are set to the value reported in Turnaev (2006) et al. [1] for the fixed-parameter model. The reaction is presumed to be at equilibrium, such that
are set to the value reported in Turnaev (2006) et al. [1] for the fixed-parameter model. The reaction is presumed to be at equilibrium, such that 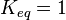 and
and 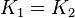 . The sampling of the parameters are done in a way so that it ranges between
. The sampling of the parameters are done in a way so that it ranges between ![[0.001\times mean \quad 1000 \times mean ]](/wiki/images/math/7/0/5/70565b13097c64cb12ee455eb2006ee7.png) to allow a large exploration of the parameter space. We use the Range rule to calculate the mean and standard deviation from maximum and minimum value. For both
to allow a large exploration of the parameter space. We use the Range rule to calculate the mean and standard deviation from maximum and minimum value. For both 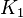 and
and 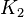 the minimum and maximum value lies between 0.0000103 and 10.3. So the mean is 5.16 and standard deviation would be 2.57499.
the minimum and maximum value lies between 0.0000103 and 10.3. So the mean is 5.16 and standard deviation would be 2.57499.
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
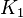
|
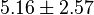
|
||
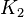
|
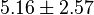
|