Difference between revisions of "Pyruvate kinase"
(→Parameters with uncertainty) |
|||
| Line 73: | Line 73: | ||
|- | |- | ||
|<math>V_{mf}</math> | |<math>V_{mf}</math> | ||
| − | |1. | + | |<math>3 \pm 1.3 (4)</math><ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi]) </ref> |
|<math> \text{mM min}^{-1} </math> | |<math> \text{mM min}^{-1} </math> | ||
|rowspan="10"|HeLa cell line | |rowspan="10"|HeLa cell line | ||
| Line 116: | Line 116: | ||
| For allosteric regulation the affinity constant is used. It is the inverted dissociation constant. so <math> Ka_{SER} = \frac{1}{K_d}</math> where <math> k_d = 0.2 mM</math> <ref name="Chaneton_2012>Chaneton, B. ''et al.''(2012) ''Serine is a natural ligand and allosteric activator of pyruvate kinase M2''. Nature 491, 458–462 </ref> | | For allosteric regulation the affinity constant is used. It is the inverted dissociation constant. so <math> Ka_{SER} = \frac{1}{K_d}</math> where <math> k_d = 0.2 mM</math> <ref name="Chaneton_2012>Chaneton, B. ''et al.''(2012) ''Serine is a natural ligand and allosteric activator of pyruvate kinase M2''. Nature 491, 458–462 </ref> | ||
|} | |} | ||
| − | |||
| − | |||
==References== | ==References== | ||
<references/> | <references/> | ||
Revision as of 14:26, 30 April 2014
Pyruvate kinase is a transferase enzyme that catalyzes the transfer of a phosphate group from phosphoenolpyruvate (PEP) to ADP, yielding one molecule of pyruvate and one molecule of ATP.
Contents
Chemical reaction
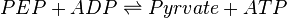
Rate equation
The rate equation is represented by the allosteric regualation model of Monod, Wyman and Changeux (MWS). Fru1,6BP and Serine are activators and ATP is inhibiting. Simple Micahelis-Menten kinetics (Briggs Haldane) is used for ADP and reverse reaction [1]
![v=V_m \left( \left(\frac{\frac{[ADP]}{K_{ADP}}}{1+\frac{[ADP]}{K_{ADP}}}\right) \left( \frac{\frac{[PEP]}{Km_{PEP}}\left( 1+\frac{[PEP]}{Km_{PEP}} \right)^3 }{ \frac{L \left( 1 + \frac{[ATP]}{Ki_{ATP}} \right)^4 }{ \left( 1 + \frac{[SER]}{Ka_{SER}} \right)^4 \left( 1 + \frac{F1,6BP}{Ka_{F1,6BP}} \right)^4 } + \left( 1 + \frac{[PEP]}{Km_{PEP}} \right)^4} \right) - \left( \frac{\frac{[ATP][PYR]}{K_{ATP} \times K_{PYR} \times K_{eq}}}{1 +\frac{[ATP]}{K_{ATP}} + \frac{[PYR]}{K_{PYR}} + \frac{[ATP][PYR]}{K_{ATP} \times K_{PYR} }} \right) \right)](/wiki/images/math/6/0/8/608585715d214881c663ba97b3810b40.png)
Parameter values
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
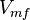
|
1.9[2] | 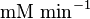
|
HeLa cell line | |
 [2] [2]
|
195172 | Recalculated from the ΔGº´ = - 31.4 KJ mol-1. | ||
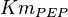 [2] [2]
|
0.014 | mM | ||
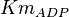 [2] [2]
|
0.4 | mM | ||
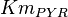 [3] [3]
|
10 | mM | ||
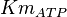 [3] [3]
|
0.86 | mM | ||
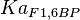 [4] [4]
|
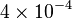
|
mM | ||
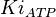 [4] [4]
|
2.5 | mM | ||
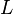 [5] [5]
|
1 | Dimensionless | ||
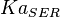
|
5 | mM | For allosteric regulation the affinity constant is used. It is the inverted dissociation constant. so 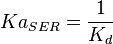 where where 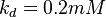 [6] [6]
|
Parameters with uncertainty
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
|
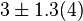 [2] [2]
|
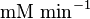
|
HeLa cell line | |
 [2] [2]
|
195172 | Recalculated from the ΔGº´ = - 31.4 KJ mol-1. | ||
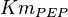 [2] [2]
|
0.014 | mM | ||
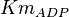 [2] [2]
|
0.4 | mM | ||
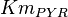 [3] [3]
|
10 | mM | ||
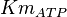 [3] [3]
|
0.86 | mM | ||
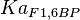 [4] [4]
|
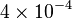
|
mM | ||
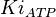 [4] [4]
|
2.5 | mM | ||
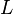 [5] [5]
|
1 | Dimensionless | ||
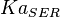
|
5 | mM | For allosteric regulation the affinity constant is used. It is the inverted dissociation constant. so 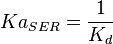 where where 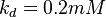 [6] [6]
|
References
- ↑ Monod J, Wyman J, Changeux J-P (1965). On the Nature of Allosteric Transitions: A Plausible Model . Journal of Molecular Biology 12:88–118 (doi)
- ↑ 2.0 2.1 2.2 2.3 2.4 2.5 2.6 2.7 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)
- ↑ 3.0 3.1 3.2 3.3 H.U. Bergmeyer. Methods of Enzymatic Analysis. Verlag Chemie, Winheim
- ↑ 4.0 4.1 4.2 4.3 Imamura K, Tanaka T (1982). Pyruvate kinase isoenzymes from rat, Methods Enzymol. 90 (1982) 150–165
- ↑ 5.0 5.1 Arbitrary value
- ↑ 6.0 6.1 Chaneton, B. et al.(2012) Serine is a natural ligand and allosteric activator of pyruvate kinase M2. Nature 491, 458–462