Difference between revisions of "Prostaglandin Transporter"
(→Parameters) |
(→Parameters) |
||
| Line 55: | Line 55: | ||
|} | |} | ||
| + | |||
| + | No data available on the turnover of PGT/ an organic molecule transporter therefore based upon estimates made by http://book.bionumbers.org/what-are-the-rates-of-membrane-transporters/ | ||
{|class="wikitable sortable" | {|class="wikitable sortable" | ||
|+ style="text-align: left;" | Turnover Number | |+ style="text-align: left;" | Turnover Number | ||
| Line 60: | Line 62: | ||
! Value | ! Value | ||
! Units | ! Units | ||
| − | ! | + | ! Transporter |
! Notes | ! Notes | ||
! Reference | ! Reference | ||
|- | |- | ||
| − | |( | + | |3.5 |
| − | | | + | |min-1 |
| − | | | + | |Glucose transporter ptsI |
| − | | | + | |Organism: Bacteria Escherichia coli |
| − | | | + | Back of the envelope calculation by BioNumbers: http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=102931 |
| + | |<ref name="Waygood1980"> [https://www.ncbi.nlm.nih.gov/pubmed/6992959?dopt=Abstract Waygood EB ''Enzyme I of the phosphoenolpyruvate: sugar phosphotransferase system of Escherichia coli. Purification to homogeneity and some properties.''Can J Biochem. 1980 Jan;58(1):40-8.]</ref> | ||
| + | |- | ||
| + | |0.33 - 0.83 | ||
| + | |min-1 | ||
| + | |Rate of transport of H+/Lactose transporter | ||
| + | |Organism: Bacteria Escherichia coli | ||
| + | Back of the envelop calculation on bionumbers http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=103159 | ||
| + | |<ref name="Naftalin2007"> [https://www.ncbi.nlm.nih.gov/pubmed/17325012?dopt=Abstract Naftalin R. J. ''ELactose permease H+-lactose symporter: mechanical switch or Brownian ratchet?''Biophys J. 2007 May 15;92(10):3474-91. Epub 2007 Feb 26.]</ref> | ||
| + | |- | ||
| + | |3.28 | ||
| + | |min-1 | ||
| + | |Catalytic Rate of transporter HXT7 | ||
| + | |Organism: Budding yeast Saccharomyces cerevisiae | ||
| + | Back of the envelope calculation http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=101737 | ||
| + | |<ref name="Ye2001"> [https://www.ncbi.nlm.nih.gov/pubmed/11561293?dopt=Abstract Ye L. ''Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiae''Yeast. 2001 Sep 30;18(13):1257-67.]</ref> | ||
|- | |- | ||
|} | |} | ||
Revision as of 11:34, 1 June 2017
Contents
Reaction
Chemical equation
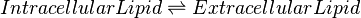
Rate equation
Parameters
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 0.0054 | mM | Human Eye | Vector: HEK-OATP2A1 or HEK-OATP2B1 and the respective vector controls
Substrate: Latanoprost, a prostaglandin F2a analogue pH: 7.5 Temp: 37 °C |
[1] |
| 0.000331 ± 0.000131 | mM | Transfected Human embryonic kidney cells (HEK293) | Substrate: PGE2
pH: unknown Temp: 37 °C |
[2] |
| 0.007202 ± 0.000595 | mM | Transfected Human embryonic kidney cells (HEK293) | Substrate: PGE3
pH: unknown Temp: 37 °C |
[2] |
| 0.000376 ± 3.4e-5 | mM | PGT-expressing Madin-Darby Canine Kidney Epithelial Cells (MDCK) cells | Substrate: PGH2
pH: unknown Temp: unknown |
[3] |
No data available on the turnover of PGT/ an organic molecule transporter therefore based upon estimates made by http://book.bionumbers.org/what-are-the-rates-of-membrane-transporters/
| Value | Units | Transporter | Notes | Reference |
|---|---|---|---|---|
| 3.5 | min-1 | Glucose transporter ptsI | Organism: Bacteria Escherichia coli
Back of the envelope calculation by BioNumbers: http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=102931 |
[4] |
| 0.33 - 0.83 | min-1 | Rate of transport of H+/Lactose transporter | Organism: Bacteria Escherichia coli
Back of the envelop calculation on bionumbers http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=103159 |
[5] |
| 3.28 | min-1 | Catalytic Rate of transporter HXT7 | Organism: Budding yeast Saccharomyces cerevisiae
Back of the envelope calculation http://bionumbers.hms.harvard.edu/bionumber.aspx?&id=101737 |
[6] |
| Value | Units | Species | Notes | Reference |
|---|---|---|---|---|
| 125 | ppm | Human | Vector:Lung
Enzyme: SLCO2A1 pH: 7.5 Temperature: 37 °C |
[7] |
| 17.4 | ppm | Human | Expression Vector: Esophagus
Enzyme: SLCO2A1 pH: 7.5 Temperature: 37 °C |
[8] |
| 2.82 | ppm | Human | Vector:Gut
Enzyme: SLCO2A1 pH: 7.5 Temperature: 37 °C |
[7] |
References
- ↑ M. Kraft The prostaglandin transporter OATP2A1 is expressed in human ocular tissues and transports the antiglaucoma prostanoid latanoprost. Invest Ophthalmol Vis Sci. 2010 May;51(5):2504-11. doi: 10.1167/iovs.09-4290. Epub 2009 Dec 17.
- ↑ 2.0 2.1 Gose T. Prostaglandin transporter (OATP2A1/SLCO2A1) contributes to local disposition of eicosapentaenoic acid-derived PGE3. Prostaglandins Other Lipid Mediat. 2016 Jan;122:10-7. doi: 10.1016/j.prostaglandins.2015.12.003. Epub 2015 Dec 10.
- ↑ Chi Y. The prostaglandin transporter PGT transports PGH2 Biochemical and Biophysical Research Communications, Volume 395, Issue 2, 30 April 2010, Pages 168–172
- ↑ Waygood EB Enzyme I of the phosphoenolpyruvate: sugar phosphotransferase system of Escherichia coli. Purification to homogeneity and some properties.Can J Biochem. 1980 Jan;58(1):40-8.
- ↑ Naftalin R. J. ELactose permease H+-lactose symporter: mechanical switch or Brownian ratchet?Biophys J. 2007 May 15;92(10):3474-91. Epub 2007 Feb 26.
- ↑ Ye L. Expression and activity of the Hxt7 high-affinity hexose transporter of Saccharomyces cerevisiaeYeast. 2001 Sep 30;18(13):1257-67.
- ↑ 7.0 7.1 M. Kim A draft map of the human proteome Nature, 2014 509, 575–581
- ↑ M. Wilhelm Mass-spectrometry-based draft of the human proteome Nature, 2014 509, 582–587