Difference between revisions of "Phosphoglycerate dehydrogenase"
(→Rate equation) |
(→Rate equation) |
||
| Line 10: | Line 10: | ||
<center><math> \frac{ serA*KcatA*\frac{[3PG]}{KA_{3PG}}}{\frac{1 + \frac{[3PG]}{KA_{3PG}} + \frac{[PHP]}{KA_{PHP}}} {1+ \frac{[SER]}{KiA_{SER}}}} </math></center> | <center><math> \frac{ serA*KcatA*\frac{[3PG]}{KA_{3PG}}}{\frac{1 + \frac{[3PG]}{KA_{3PG}} + \frac{[PHP]}{KA_{PHP}}} {1+ \frac{[SER]}{KiA_{SER}}}} </math></center> | ||
| − | <center><math> \frac{\left( | + | <center><math> \frac{\left( \frac{serA*KcatA*\frac{[3PG]}{KA_{3PG}}} {} \right)}{} </math></center> |
==Parameters== | ==Parameters== | ||
Revision as of 16:16, 30 July 2014
The enzyme Phosphoglycerate dehydrogenase (PDH) catalyzes the reaction to convert 3-phospho-D-glycerate (P3G) to phosphonooxypyruvate (PHP). The cofactors NADH and NAD+.
Contents
Chemical equation
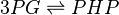
Rate equation
Modified rate equation is used [1]
Parameters
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
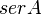
|
1.15 [2] | mM | Escherichia coli | |
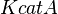
|
0.55 [3] | 1/s | ||
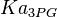
|
1.2[3] | mM | ||
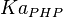
|
0.0032[3] | mM | ||
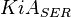
|
0.0038[3] | mM |
Parameters with uncertainty
- serA is the enzyme concentration collected from Turnaev et. al. [2] which is considered to be fixed.
- Data reported in Zhao et. al. [3] mentioned that the Standard Errors of parameters were less than 10% in their data. For the sake of completeness we consider the maximum Standard Error of 10% for all the data. Considering this the Standard Error for
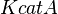 would be
would be 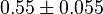 . Considering three samples for the data the standard deviation for this parameter would be 0.09 (Std. Dev = Std. Error
. Considering three samples for the data the standard deviation for this parameter would be 0.09 (Std. Dev = Std. Error 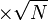 ).
). - For
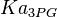 it would be
it would be 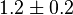 , for
, for 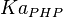 it is
it is 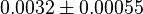 and for
and for 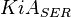 it is
it is 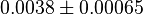
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
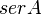
|
1.15 [2] | mM | Escherichia coli | |
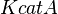
|
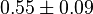
|
1/s | ||
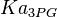
|
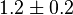
|
mM | ||
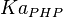
|
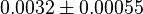
|
mM | ||
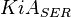
|
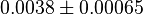
|
mM |
References
- ↑ Smallbone K, Stanford NJ (2013). Kinetic modeling of metabolic pathways: Application to serine biosynthesis. In: Systems Metabolic Engineering, Humana Press. pp. 113–121
- ↑ 2.0 2.1 2.2 Turnaev II, Ibragimova SS, Usuda Y et al (2006). Mathematical modeling of serine and glycine synthesis regulation in Escherichia coli. Proceedings of the fifth international conference on bioinformatics of genome regulation and structure 2:78–83
- ↑ 3.0 3.1 3.2 3.3 3.4 Zhao G, Winkler ME (1996). A novel alphaketoglutarate reductase activity of the serA-encoded 3-phosphoglycerate dehydrogenase of Escherichia coli K-12 and its possible implications for human 2-hydroxyglutaric aciduria. J Bacteriol 178:232–239