Difference between revisions of "Nucleosid diphosphate kinase"
(→Parameters with uncertainty) |
|||
| Line 60: | Line 60: | ||
* Two values of <math>V_{max}</math> is also reported in Fukuchi (1994) et. al. <ref name="Fukuchi_1994"></ref>; 126 (4.20 <math>\text{mmol min}^{-1}\text{ mg}^{-1}</math> multiplied with 30 mg) and 138 (4.60 <math>\text{mmol min}^{-1}\text{ mg}^{-1}</math> multiplied with 30 mg) <math>\text{mmol min}^{-1}</math>. Taking average gives <math>132 \pm 8.48</math>. | * Two values of <math>V_{max}</math> is also reported in Fukuchi (1994) et. al. <ref name="Fukuchi_1994"></ref>; 126 (4.20 <math>\text{mmol min}^{-1}\text{ mg}^{-1}</math> multiplied with 30 mg) and 138 (4.60 <math>\text{mmol min}^{-1}\text{ mg}^{-1}</math> multiplied with 30 mg) <math>\text{mmol min}^{-1}</math>. Taking average gives <math>132 \pm 8.48</math>. | ||
| − | *Standard free energy <math>\Delta G^{o}{'}</math> is reported to be zero kJ/mol. Therefore the value of Keq | + | *Standard free energy <math>\Delta G^{o}{'}</math> is reported to be zero kJ/mol. Therefore the value of Keq can be assumed to be constant. As there was no reported other value std. dev. from other Keq values are considered which is <math>\approx 30%</math>. So the Keq value would be <math>1 \pm 0.3</math> |
{|class="wikitable" | {|class="wikitable" | ||
| Line 92: | Line 92: | ||
|- | |- | ||
|<math>K_{eq}</math> | |<math>K_{eq}</math> | ||
| − | |<math>1</math> | + | |<math>1 \pm .3</math> |
|Dimensionless | |Dimensionless | ||
|} | |} | ||
Revision as of 15:49, 28 May 2014
Nucleoside-diphosphate kinases are enzymes that catalyze the exchange of phosphate groups between different nucleotides. The overall effect of NDKs is to transfer a phosphate group from a nucleoside triphosphate to a nucleoside diphosphate. Starting with ATP and UDP, the activity of NDK produces ADP and UTP.
Contents
Chemical equation
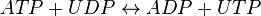
Rate equation
Random order Bi-Bi rate law is used from [1]
![\frac{ \frac{V_{max}}{K_m^{ATP}K_m^{UDP}}\left( [ATP][UDP] - \frac{[ADP][UTP]}{K_{eq}} \right) } { \left( 1 + \frac{[ATP]}{K_{m}^{ATP}} \right)\left( 1 + \frac{[UDP]}{K_{m}^{UDP}} \right) + \left( 1 + \frac{[ADP]}{K_{m}^{ADP}} \right)\left( 1 + \frac{[UTP]}{K_{m}^{UTP}} \right) -1 }](/wiki/images/math/9/e/6/9e6a0b14129bad329a3c09d3c7a69f43.png)
Parameter values
- A mean body weight is defined as 75Kg in Konig et. al [1]. The
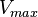 is calculated by multiplying the reported value with body weight.
is calculated by multiplying the reported value with body weight.
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
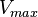
|
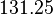 [2] [2]
|

|
Human Liver | |
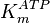
|
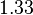 [3] [3]
|
mM | Rat liver | |
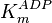
|
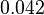 [3] [3]
|
mM | Rat liver | |
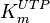
|
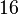 [4] [4]
|
mM | Rat tissue | |
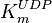
|
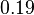 [3] [3]
|
mM | Rat liver | |

|
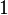
|
Dimensionless |
Parameters with uncertainty
- In the paper by Walter (1980) et. al. [3] two values of
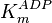 ,
, 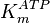 and
and 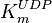 have been reported; one for cytosolic kinase and the other for membrane-associated kinase. As in our model we are not differentiating between cytosolic and membrane-associated kinase, we would consider both those values and calculate the uncertainty from them.
have been reported; one for cytosolic kinase and the other for membrane-associated kinase. As in our model we are not differentiating between cytosolic and membrane-associated kinase, we would consider both those values and calculate the uncertainty from them.
- Similarly two values of
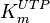 has been reported in Fukuchi (1994) et. al. [4]; for
has been reported in Fukuchi (1994) et. al. [4]; for 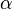 kinase and
kinase and 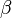 kinase. Taking average of those kinase gives a value of
kinase. Taking average of those kinase gives a value of 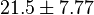
- Two values of
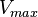 is also reported in Fukuchi (1994) et. al. [4]; 126 (4.20
is also reported in Fukuchi (1994) et. al. [4]; 126 (4.20 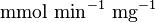 multiplied with 30 mg) and 138 (4.60
multiplied with 30 mg) and 138 (4.60 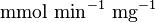 multiplied with 30 mg)
multiplied with 30 mg) 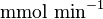 . Taking average gives
. Taking average gives 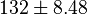 .
.
- Standard free energy
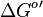 is reported to be zero kJ/mol. Therefore the value of Keq can be assumed to be constant. As there was no reported other value std. dev. from other Keq values are considered which is Failed to parse (Cannot store math image on filesystem.): \approx 30%
. So the Keq value would be Failed to parse (Cannot store math image on filesystem.): 1 \pm 0.3
is reported to be zero kJ/mol. Therefore the value of Keq can be assumed to be constant. As there was no reported other value std. dev. from other Keq values are considered which is Failed to parse (Cannot store math image on filesystem.): \approx 30%
. So the Keq value would be Failed to parse (Cannot store math image on filesystem.): 1 \pm 0.3
| Parameter | Value | Units | Organism | Remarks |
|---|---|---|---|---|
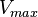
|
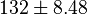
|

|
Rat lever | |
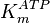
|
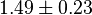
|
mM | ||
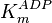
|
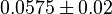
|
mM | ||
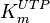
|
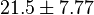
|
mM | ||
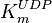
|
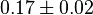
|
mM | ||

|
Failed to parse (Cannot store math image on filesystem.): 1 \pm .3 | Dimensionless |
References
- ↑ 1.0 1.1 M. König, S. Bulik, H.G. Holzhütter (2012), Quantifying the contribution of the liver to glucose homeostasis: a detailed kinetic model of human hepatic glucose metabolism, PLoS Comput. Biol., 8 (6), p. e1002577
- ↑ Villar-Palasi C & Larner J (1960). Levels of activity of the enzymes of the glycogen cycle in rat tissues. Arch Biochem Biophys 86, 270–273.
- ↑ 3.0 3.1 3.2 3.3 Walter P and Blobel G. (1980), Purification of a membrane-associated protein complex required for protein translocation across the endoplasmic reticulum, Proc. Natl. Acad. Sci. 77:7112-7116
- ↑ 4.0 4.1 4.2 Fukuchi, T., Shimada, N., Hanai, N., Ishikawa, N., Watanabe, K., and Kimura, N. (1994) Recombinant rat nucleoside diphosphate kinase isoforms (
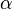 and
and 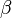 ): purification, properties and application to immunological detection of native isoforms in rat tissues. Biochim. Biophys. Acta 1205, 113–122
): purification, properties and application to immunological detection of native isoforms in rat tissues. Biochim. Biophys. Acta 1205, 113–122