Mitocondrial pyruvate metabolism
Mitocondrial pyruvate metabolism(MPM) is an enzyme that generates ATP form pyruvate.
Chemical reaction
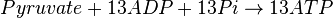
Rate equation
- Chemical reactions proceed to equilibrium within closed systems. For a simple reaction
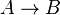 it is defined as
it is defined as ![K_{eq} = \frac{[B]_{eq}}{[A]_{eq}}](/wiki/images/math/a/c/8/ac844b70f8f7c5f1d91accb00061a1ea.png) where forward and reverse rates are equal.
where forward and reverse rates are equal. - Equilibrium is not reached in open system due to influx and outflux. Mass action ratio[1]
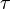 for
for 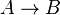 reaction is defined as
reaction is defined as ![\tau = \frac{[B]_{ob}}{[A]_{ob}}](/wiki/images/math/8/0/5/8050a810298db95df036d87c2e79f1ea.png) where subscript ob represents observable at a given point.
where subscript ob represents observable at a given point. - Deviation from equilibrium is measured with Disequilibrium constant
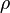 as
as 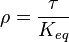
- Given the simple uni molecular reaction
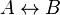 the mass action equation can be modified as
the mass action equation can be modified as
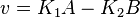
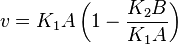
Considering 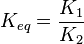 and
and 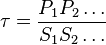 we have,
we have,
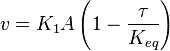
- The generalized arbitrary mass action ratio gives us
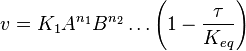
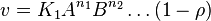
For eg. for the reaction 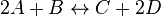 , the rate law would be
, the rate law would be 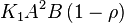
- This equation demonstrates how a rate expression can be divided into parts that include both kinetics and thermodynamic properties [2].
- Given the net rate of reaction
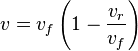 , we have
, we have
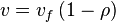
In this model
- The rate law is defined as
![v = K_1[Pyruvate][ADP]^{13}[Pi]^{13}\left(1-\frac{\frac{[ATP]^{13}}{[Pyruvate][ADP]^{13}[Pi]^{13}}}{K_{eq}}\right)](/wiki/images/math/c/0/7/c07d97f8c92fac816401bb1210a69046.png)
- The
 value for the reactions that converts pyruvate has been defined as Failed to parse (Cannot store math image on filesystem.): 3.32e^5
in [3]
value for the reactions that converts pyruvate has been defined as Failed to parse (Cannot store math image on filesystem.): 3.32e^5
in [3]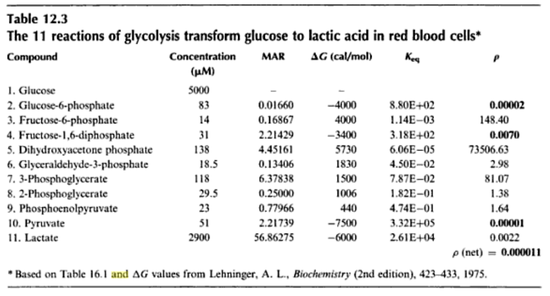
- The flux value at steady state is
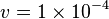 [4].
[4]. - The steady state concentrations for substrates and products are
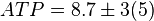 ,
, 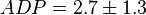 ,
, 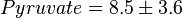 and
and 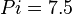 .
. - The
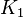 value calculated from the above mentioned values are
value calculated from the above mentioned values are
Parameter values
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
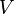
|
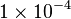 [4] [4]
|
HeLa cell line | Constant flux |
References
- ↑ Hess B. and Brand K. (1965), Enzymes and metabolite profiles. In Control of energy metabolism. III. Ed. B. Chance, R. K. Estabrook and J. R. Williamson. New York: Academic Press
- ↑ Sauro H M, Enzyme Kinetics for Systems Biology, Second Edition, Ambrosius Publishing (2013), ISBN-10: 0-9824773-3-3
- ↑ Owusu-Apenten R. Introduction to Food Chemistry, First Edition, CRC Press (2004), ISBN-10: 084931724X
- ↑ 4.0 4.1 Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)