Difference between revisions of "Mitocondrial pyruvate metabolism"
(→In this model) |
|||
| Line 29: | Line 29: | ||
*The rate law is defined as <center><math>v = K_1[Pyruvate][ADP]^{13}[Pi]^{13}\left(1-\frac{\frac{[ATP]^{13}}{[Pyruvate][ADP]^{13}[Pi]^{13}}}{K_{eq}}\right)</math></center><br> | *The rate law is defined as <center><math>v = K_1[Pyruvate][ADP]^{13}[Pi]^{13}\left(1-\frac{\frac{[ATP]^{13}}{[Pyruvate][ADP]^{13}[Pi]^{13}}}{K_{eq}}\right)</math></center><br> | ||
*The overall standard free-energy change for Pyruvate metabolism is <math>\Delta G^o{'}= -50.3 Kj/Mol</math><ref>Nelson D. and Cox M. (2008), ''Lehninger Principles of Biochemistry'', Fight Edition, W.H. Freeman and Company, ISBN-10: 071677108X</ref><ref name="Takusagawas_Note">http://crystal.res.ku.edu/taksnotes/Biol_638/notes/chp_16.pdf</ref>. '''Note''' The overall standard free energy is calculated by adding the standard free energy for all the reactions in TCA cycle. | *The overall standard free-energy change for Pyruvate metabolism is <math>\Delta G^o{'}= -50.3 Kj/Mol</math><ref>Nelson D. and Cox M. (2008), ''Lehninger Principles of Biochemistry'', Fight Edition, W.H. Freeman and Company, ISBN-10: 071677108X</ref><ref name="Takusagawas_Note">http://crystal.res.ku.edu/taksnotes/Biol_638/notes/chp_16.pdf</ref>. '''Note''' The overall standard free energy is calculated by adding the standard free energy for all the reactions in TCA cycle. | ||
| − | ::Calculating <math>K_{eq}</math> value from these free energy gives <math>\Delta G' = - 50.3\ kJ.mol^{-1}</math>, <math>Keq = exp(-\frac{\Delta G'}{RT}) = exp(\frac{50300}{8.31*298.15}) \approx | + | ::Calculating <math>K_{eq}</math> value from these free energy gives <math>\Delta G' = - 50.3\ kJ.mol^{-1}</math>, <math>Keq = exp(-\frac{\Delta G'}{RT}) = exp(\frac{50300}{8.31*298.15}) \approx 656010875.924588</math>. In order to ensure that the uncertainty does not affect the model equilibrium a small uncertainty of 5% can be considered for transporter. In our model we have applied this approach. So <math>K_{eq} = 654904512.15 \pm 32800543.79</math>. |
*The Flux of pyruvate consumed by mitochondria measured for AS_30D is <math> v = 1.8</math> <ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi])</ref>. | *The Flux of pyruvate consumed by mitochondria measured for AS_30D is <math> v = 1.8</math> <ref name="Hernandez2011"> Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 ([http://dx.doi.org/10.1016/j.bbabio.2010.11.006 doi])</ref>. | ||
*The initial concentrations for substrates and products are <math>ATP=8.7 \pm 3 (5)</math>, <math>ADP = 2.7 \pm 1.3</math>, <math>Pyruvate = 8.5 \pm 3.6</math> and <math>Pi = 7.5</math>. | *The initial concentrations for substrates and products are <math>ATP=8.7 \pm 3 (5)</math>, <math>ADP = 2.7 \pm 1.3</math>, <math>Pyruvate = 8.5 \pm 3.6</math> and <math>Pi = 7.5</math>. | ||
| − | * | + | * Considering the stoichiometry of the model the value of <math>K_1</math> is <math>5.1344e-017</math>. If we ingnore the stiochiometric constants while calculating <math>K_1</math>, the value calculated from the above mentioned values are <math>0.01045</math> |
*To calculate the uncertainty of <math>K_1</math> we first looked at the uncertainty on the substrate and product concentration. The maximum uncertainty reported for these values are <math>\approx 50%</math>. In our model we considered this <math>50%</math> uncertainty in its mean value giving value of <math>0.01045 \pm 0.0052</math> | *To calculate the uncertainty of <math>K_1</math> we first looked at the uncertainty on the substrate and product concentration. The maximum uncertainty reported for these values are <math>\approx 50%</math>. In our model we considered this <math>50%</math> uncertainty in its mean value giving value of <math>0.01045 \pm 0.0052</math> | ||
Latest revision as of 13:20, 12 August 2014
Mitocondrial pyruvate metabolism(MPM) is a pseudo reaction that represents the total ATP production from one unit of pyruvate in the mitochondrian.
Contents
Chemical reaction
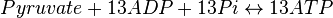
Rate equation
- Chemical reactions proceed to equilibrium within closed systems. For a simple reaction
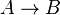 it is defined as
it is defined as ![K_{eq} = \frac{[B]_{eq}}{[A]_{eq}}](/wiki/images/math/a/c/8/ac844b70f8f7c5f1d91accb00061a1ea.png) where forward and reverse rates are equal.
where forward and reverse rates are equal. - Equilibrium is not reached in open system due to influx and outflux. Mass action ratio[1]
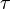 for
for 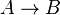 reaction is defined as
reaction is defined as ![\tau = \frac{[B]_{ob}}{[A]_{ob}}](/wiki/images/math/8/0/5/8050a810298db95df036d87c2e79f1ea.png) where subscript ob represents observable at a given point.
where subscript ob represents observable at a given point. - Deviation from equilibrium is measured with Disequilibrium constant
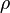 as
as 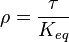
- Given the simple uni molecular reaction
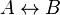 the mass action equation can be modified as
the mass action equation can be modified as
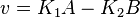
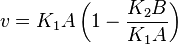
Considering 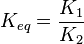 and
and 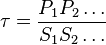 we have,
we have,
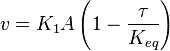
- The generalized arbitrary mass action ratio gives us
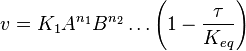
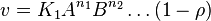
For eg. for the reaction 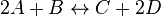 , the rate law would be
, the rate law would be 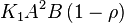
- This equation demonstrates how a rate expression can be divided into parts that include both kinetics and thermodynamic properties [2].
- Given the net rate of reaction
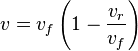 , we have
, we have
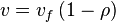
In this model
- The rate law is defined as
![v = K_1[Pyruvate][ADP]^{13}[Pi]^{13}\left(1-\frac{\frac{[ATP]^{13}}{[Pyruvate][ADP]^{13}[Pi]^{13}}}{K_{eq}}\right)](/wiki/images/math/c/0/7/c07d97f8c92fac816401bb1210a69046.png)
- The overall standard free-energy change for Pyruvate metabolism is
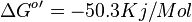 [3][4]. Note The overall standard free energy is calculated by adding the standard free energy for all the reactions in TCA cycle.
[3][4]. Note The overall standard free energy is calculated by adding the standard free energy for all the reactions in TCA cycle.
- Calculating
 value from these free energy gives
value from these free energy gives 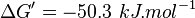 ,
, 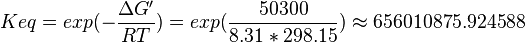 . In order to ensure that the uncertainty does not affect the model equilibrium a small uncertainty of 5% can be considered for transporter. In our model we have applied this approach. So
. In order to ensure that the uncertainty does not affect the model equilibrium a small uncertainty of 5% can be considered for transporter. In our model we have applied this approach. So 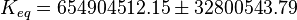 .
.
- Calculating
- The Flux of pyruvate consumed by mitochondria measured for AS_30D is
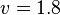 [5].
[5]. - The initial concentrations for substrates and products are
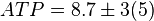 ,
, 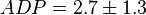 ,
, 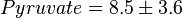 and
and 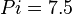 .
. - Considering the stoichiometry of the model the value of
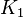 is
is 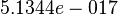 . If we ingnore the stiochiometric constants while calculating
. If we ingnore the stiochiometric constants while calculating 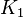 , the value calculated from the above mentioned values are
, the value calculated from the above mentioned values are 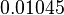
- To calculate the uncertainty of
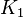 we first looked at the uncertainty on the substrate and product concentration. The maximum uncertainty reported for these values are
we first looked at the uncertainty on the substrate and product concentration. The maximum uncertainty reported for these values are 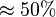 . In our model we considered this
. In our model we considered this 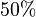 uncertainty in its mean value giving value of
uncertainty in its mean value giving value of 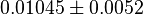
- Alternative As this is a psuedo-reaction, the above mentioned formula fails to reach steady state. So a constant flux mentioned in Hernandez et. al. can be used. The maximum relative precent of error for other constant fluxes are reported to be 31%. We consider the same relative percent error for this reaction because of it being a psuedo-reaction.
Parameter values
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
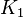
|
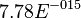 In our model we used In our model we used 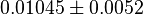
|
||

|
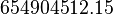
|
Alternative
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
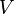
|
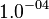
|
Constant Flux |
Parameters with uncertainty
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
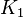
|
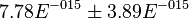
|
||

|
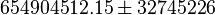
|
Alternative
| Parameter | Value | Organism | Remarks |
|---|---|---|---|
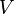
|
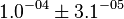
|
Constant Flux |
References
- ↑ Hess B. and Brand K. (1965), Enzymes and metabolite profiles. In Control of energy metabolism. III. Ed. B. Chance, R. K. Estabrook and J. R. Williamson. New York: Academic Press
- ↑ Sauro H M, Enzyme Kinetics for Systems Biology, Second Edition, Ambrosius Publishing (2013), ISBN-10: 0-9824773-3-3
- ↑ Nelson D. and Cox M. (2008), Lehninger Principles of Biochemistry, Fight Edition, W.H. Freeman and Company, ISBN-10: 071677108X
- ↑ http://crystal.res.ku.edu/taksnotes/Biol_638/notes/chp_16.pdf
- ↑ Marín-Hernández A, Gallardo-Pérez JC, Rodríguez-Enríquez S et al (2011) Modeling cancer glycolysis. Biochim Biophys Acta 1807:755–767 (doi)