Mevalonate pathway with limonene synthesis
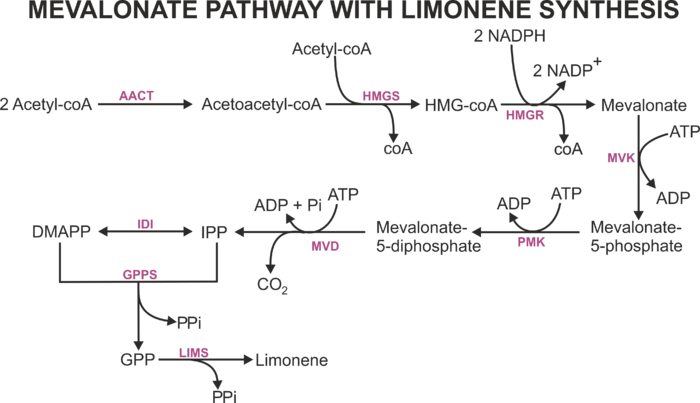
The mevalonate (MVA) pathway is one of two independent pathways that is responsible to produce isopentenyl diphosphate (IPP) and dimethylallyl diphosphate (DMAPP) -- the two main precursors to produce terpenes. This pathway has seven enzymes to produce IPP and DMAPP, and these compounds are consumed to form geranyl diphosphate (GPP) -- the precursor for the synthesis of monoterpenes such as limonene.
Contents
Mevalonate pathway models
We have two MVA pathway models. Both models describe the synthesis of limonene, but one is more detailed than the other. In the basic MVA model, the enzyme concentrations are set as fixed concentrations whereas in the improved MVA model, the enzyme concentrations are not fixed and its degradation is taken into account. A detailed description of these models along with the simulations and analysis that were carried out can be accessed by clicking on the buttons below.
Common Mevalonate pathway reactions (Reactions in all versions mevalonate models)
Parameterisation
Parameterisation for all of the MVA models here are according to our DIPPER protocol with the DipperXBrenda add-on.
Weighting scheme used
| Categories | Subcategories | Description | Weight value |
|---|---|---|---|
| Method | In vivo | Ideal parameter value measurement condition (i.e. measurement condition matches the modelling system) | 4 |
| In vitro | Parameter value was measured in vitro, and does not match our modelling system but the values are still useful. | 1 | |
| Organism | Identical | The ideal organism source where the enzyme is characterised from is E. coli, which the organism being modelled in this example. | 4 |
| Related | The organism from which the enzyme is characterised is an organism which is phylogenetically related to E. coli | 2 | |
| Unrelated | The organism from which the enzyme is characterised is not E. coli and not phylogenetically related to E. coli. | 1 | |
| Enzyme class | Identical | Ideal EC would be EC 4.2.3.16, which is the exact EC for limonene synthase. | 4 |
| Related | This is chosen when the parameter value is found for the same class or subclass as the modelled enzyme. In this case study example, values from EC 4.*.*.* or EC 4.3.2.* would be assigned to this weighting subcategory. | 2 | |
| Different | This is for parameter values originating from enzymes with a completely different enzyme class. The value would still be useful in cases where parameter values for a certain enzyme is hard to find and the modeller can look for information from other enzymes (e.g. there are values found for the kinetics of a substrate of interest but with a different enzyme). | 1 | |
| pH | Identical | Ideal pH that matches the modelling system. In this case study example, it is pH 7.5. | 4 |
| Similar | This is chosen for when the pH determined during kinetic evaluation is close to our ideal pH. In this case study example, we deemed a pH value within ± 0.5 from the ideal 7.5 as similar (i.e. pH 7.0 and pH 8.0). | 2 | |
| Different | This is chosen when the pH is different from the two subcategories described above. | 1 | |
| Temperature | Identical | Ideal temperature that matches the modelling system. In this case study example, the ideal temperature is 30°C. | 4 |
| Similar | As in the same subcategory for pH, this is chosen when the temperature is within ±0.5 from the ideal value (i.e. 25°C and 35°C) | 2 | |
| Different | This is chosen when the temperature during measurement is different from the two subcategories described above. | 1 |

