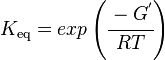Difference between revisions of "Menthol:neomenthol reductase (MNMR)"
Aliah.hawari (talk | contribs) (→Published Kinetic Parameter Values) |
Aliah.hawari (talk | contribs) (→Km Values) |
||
| Line 230: | Line 230: | ||
=== Published Kinetic Parameter Values === | === Published Kinetic Parameter Values === | ||
| − | ==== Km Values ==== | + | ==== Km Values for menthone, isomenthone, neomenthol, neoisomenthol and NADPH ==== |
| − | {| class="wikitable" | + | {| class="wikitable sortable" |
! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 50px;" | Parameter | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 50px;" | Parameter | ||
! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 100px;" | Directionality | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 100px;" | Directionality | ||
| Line 240: | Line 240: | ||
! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 10px;" | References | ! style="border: 1px solid black; padding: 5px; background: #ffdead; width: 10px;" | References | ||
|- | |- | ||
| + | | Km || Forward || menthone || 96 || µM || Km: 96.0 +/- 5.2. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 101.2 || µM || Km: 96.0 +/- 5.2. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 90.8 || µM || Km: 96.0 +/- 5.2. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Kcat || Forward || menthone || 0.91 || 1/s || Kcat: 0.89 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Kcat || Forward || menthone || 0.87 || 1/s || Kcat: 0.89 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Kcat || Forward || isomenthone || 0.44 || 1/s || Kcat: 0.44 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Kcat || Forward || isomenthone || 0.46 || 1/s || Kcat: 0.44 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Kcat || Forward || isomenthone || 0.42 || 1/s || Kcat: 0.44 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Km || Forward || isomenthone || 186.5 || µM || Km: 186.5 +/- 16.8. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Km || Forward || isomenthone || 203.3 || µM || Km: 186.5 +/- 16.8. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Km || Forward || isomenthone || 169.7 || µM || Km: 186.5 +/- 16.8. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING || Toogood2016 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 7.1 || µM || producing neomenthol. KM: 7.1 +/- 1.4. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 8.5 || µM || producing neomenthol. KM: 7.1 +/- 1.4. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 5.7 || µM || producing neomenthol. KM: 7.1 +/- 1.4. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Km || Reverse || neomenthol || 305 || µM || producing menthone from neomenthol (reverse). KM: 305 +/- 100. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Km || Reverse || neomenthol || 405 || µM || producing menthone from neomenthol (reverse). KM: 305 +/- 100. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Km || Reverse || neomenthol || 205 || µM || producing menthone from neomenthol (reverse). KM: 305 +/- 100. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Kcat || Forward || menthone || 0.6 || 1/s || producing neomenthol (reverse). Kcat: 0.60. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Kcat || Reverse || neomenthol || 0.91 || 1/s || producing neomenthol (reverse). Kcat: 0.91. from Artemisia annua, pH 7.0, Temp 30C, || Ryden2010 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 674 || µM || menthone -> neomenthone, Km: 674 +/- 78, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 752 || µM || menthone -> neomenthone, Km: 674 +/- 78, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 596 || µM || menthone -> neomenthone, Km: 674 +/- 78, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || isomenthone || 1000 || µM || isomenthone -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || NADPH || 10 || µM || isomenthone/menthone + nadph -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || NADPH || 11 || µM || isomenthone/menthone + nadph -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || NADPH || 9 || µM || isomenthone/menthone + nadph -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. || Davis2005 | ||
| + | |- | ||
| + | | Km || Forward || menthone || 22 || µM || menthone -> neomenthol, pH 7.5, 30C, from Mentha piperita, expressed in E. coli || Kjonaas1982 | ||
| + | |- | ||
| + | | Km || Forward || NADPH || 20 || µM || menthone -> neomenthol, pH 7.5, 30C, from Mentha piperita, expressed in E. coli || Kjonaas1982 | ||
|} | |} | ||
Latest revision as of 11:48, 3 June 2016
You can go back to main page of the kinetic model here.
Legend:
| Have not started ·· | 1 -2 data found ·· | 3-4 data found ·· | sufficient data found/estimated ·· | data distribution generated ·· | data sampled |
| to do | DONE! |
Contents
- 1 What we know
- 2 Reaction catalysed
- 3 Enzyme and Metabolite Background Information
- 4 Equation Rate
- 5 Strategies for estimating the kinetic parameter values
- 6 Simulations
- 7 References
What we know
Menthone: neomenthol reductase(s) (MNMR) catalyses the NADPH-dependent convertion of menthone to neomenthol and the conversion of isomenthone to neoisomenthol.
Issues
Strategies
Reaction catalysed
Enzyme and Metabolite Background Information
Long metabolite names are abbreviated in the model for clarity and standard identification purposes.
| Metabolite | Abbreviation | Chemical Formula | Molar mass (g/mol) | ChEBI | ChEMBL | PubChem | BRENDA | PlantCyc |
|---|---|---|---|---|---|---|---|---|
| menthone:menthol reductase | MMR | 34070 Da [1], 35000 Da [2] | 1.1.1.207 | |||||
| menthone | ||||||||
| isomenthone | ||||||||
| NADPH | C21H30N7O17P3 | 745.42116 | 16474 | |||||
| NADP+ | C21H29N7O17P3 | 744.41322 | 18009 |
Equation Rate
Two MMR reactions are included in the kinetic model with one converting menthone to neomenthol, and one converting isomenthone to neoisomenthol. Both reactions are parameterised using random Bi-Bi reversible Michaelis-Menten equation.
Reaction 1: Conversion of menthone to neomenthol
Reaction 2: Conversion of isomenthone to neoisomenthol
| Parameter | Description | Units |
|---|---|---|
| VMMR | Reaction rate for MNMR | μM/min |
| Kcatforward | Catalytic constant in the forward direction | s-1 |
| Kmmenthone | Michaelis-Menten constant for menthone | μM |
| Kmneomenthol | Michaelis-Menten constant for neomenthol | μM |
| Kmisomenthone | Michaelis-Menten constant for isomenthone | μM |
| Kmneoisomenthol | Michaelis-Menten constant for neoisomenthol | μM |
| KmNADPH | Michaelis-Menten constant for NADPH | μM |
| KmNADP | Michaelis-Menten constant for NADP+ | μM |
| Keq | Equilibrium constant | |
| [MMR] | enzyme concentration | μM |
| [neomenthol] | neomenthol concentration | μM |
| [menthone] | Menthone concentration | μM |
| [isomenthone] | Isomenthone concentration | μM |
| [neoisomenthol] | neoisomenthol concentration | μM |
| [NADPH] | NADPH concentration | μM |
| [NADP] | NADP+ concentration | μM |
Strategies for estimating the kinetic parameter values
Standard Gibbs Free energy
The Gibbs free energy for MMR is -1.6264343 kcal.mol-1. This value is estimated from the 'Contribution group' method by Latendresse, M. and is available from MetaCyc (EC 1.1.1.207) [3].
Calculating the Equilibrium Constant
The equilibrium constant can be calculated using the Van't Hoff Isotherm equation:
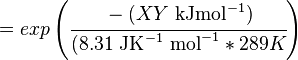
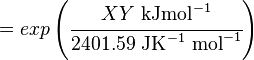
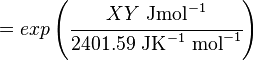
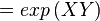
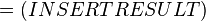
where;
| Keq | Equilibrium constant |
| -ΔG° | Gibbs free energy change. For (INSERT ENZYME) it is (INSERT VALUE) kJmol-1 |
| R | Gas constant with a value of 8.31 JK-1mol-1 |
| T | Temperature which is always expressed in kelvin |
Extracting Information from menthone Production Rates
A table will go here
Published Kinetic Parameter Values
Km Values for menthone, isomenthone, neomenthol, neoisomenthol and NADPH
| Parameter | Directionality | Substrate / Product | Value | unit | Method notes | References |
|---|---|---|---|---|---|---|
| Km | Forward | menthone | 96 | µM | Km: 96.0 +/- 5.2. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Km | Forward | menthone | 101.2 | µM | Km: 96.0 +/- 5.2. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Km | Forward | menthone | 90.8 | µM | Km: 96.0 +/- 5.2. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Kcat | Forward | menthone | 0.91 | 1/s | Kcat: 0.89 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Kcat | Forward | menthone | 0.87 | 1/s | Kcat: 0.89 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Kcat | Forward | isomenthone | 0.44 | 1/s | Kcat: 0.44 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Kcat | Forward | isomenthone | 0.46 | 1/s | Kcat: 0.44 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Kcat | Forward | isomenthone | 0.42 | 1/s | Kcat: 0.44 +/- 0.02. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Km | Forward | isomenthone | 186.5 | µM | Km: 186.5 +/- 16.8. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Km | Forward | isomenthone | 203.3 | µM | Km: 186.5 +/- 16.8. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Km | Forward | isomenthone | 169.7 | µM | Km: 186.5 +/- 16.8. From Helen's data, would need to ask for experimental conditions, unpublished. Assume optimal conditions. NEEDS CONFIRMING | Toogood2016 |
| Km | Forward | menthone | 7.1 | µM | producing neomenthol. KM: 7.1 +/- 1.4. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Km | Forward | menthone | 8.5 | µM | producing neomenthol. KM: 7.1 +/- 1.4. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Km | Forward | menthone | 5.7 | µM | producing neomenthol. KM: 7.1 +/- 1.4. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Km | Reverse | neomenthol | 305 | µM | producing menthone from neomenthol (reverse). KM: 305 +/- 100. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Km | Reverse | neomenthol | 405 | µM | producing menthone from neomenthol (reverse). KM: 305 +/- 100. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Km | Reverse | neomenthol | 205 | µM | producing menthone from neomenthol (reverse). KM: 305 +/- 100. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Kcat | Forward | menthone | 0.6 | 1/s | producing neomenthol (reverse). Kcat: 0.60. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Kcat | Reverse | neomenthol | 0.91 | 1/s | producing neomenthol (reverse). Kcat: 0.91. from Artemisia annua, pH 7.0, Temp 30C, | Ryden2010 |
| Km | Forward | menthone | 674 | µM | menthone -> neomenthone, Km: 674 +/- 78, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | menthone | 752 | µM | menthone -> neomenthone, Km: 674 +/- 78, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | menthone | 596 | µM | menthone -> neomenthone, Km: 674 +/- 78, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | isomenthone | 1000 | µM | isomenthone -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | NADPH | 10 | µM | isomenthone/menthone + nadph -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | NADPH | 11 | µM | isomenthone/menthone + nadph -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | NADPH | 9 | µM | isomenthone/menthone + nadph -> neomenthone, Km: >1000, pH 9.3, temp 30 c, from Mentha piperita, expressed in E.coli. | Davis2005 |
| Km | Forward | menthone | 22 | µM | menthone -> neomenthol, pH 7.5, 30C, from Mentha piperita, expressed in E. coli | Kjonaas1982 |
| Km | Forward | NADPH | 20 | µM | menthone -> neomenthol, pH 7.5, 30C, from Mentha piperita, expressed in E. coli | Kjonaas1982 |
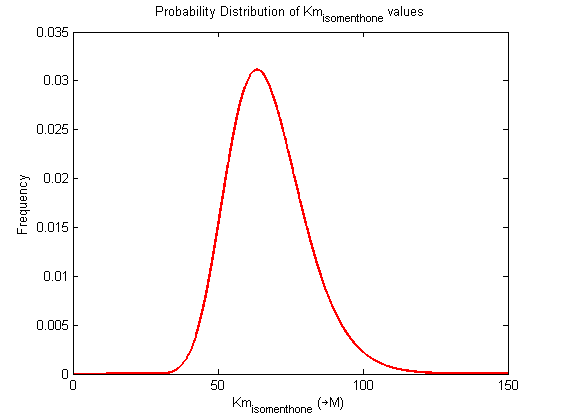
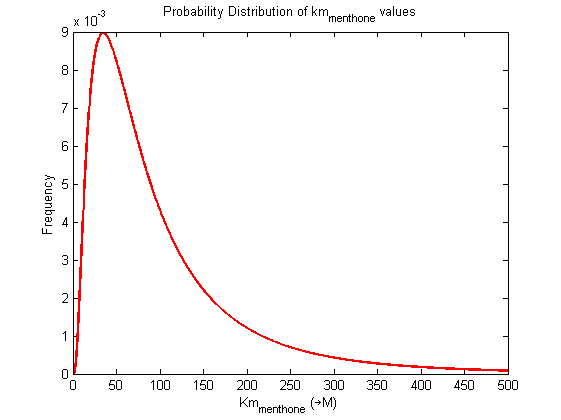
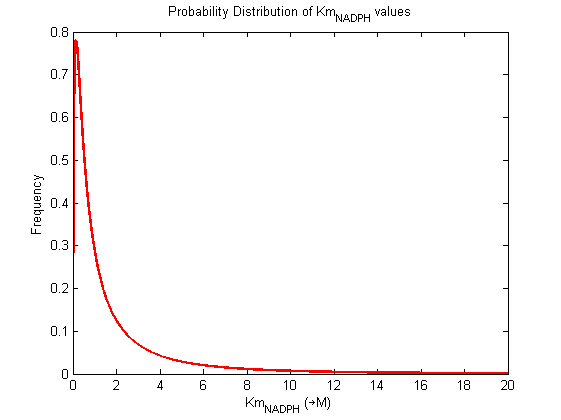
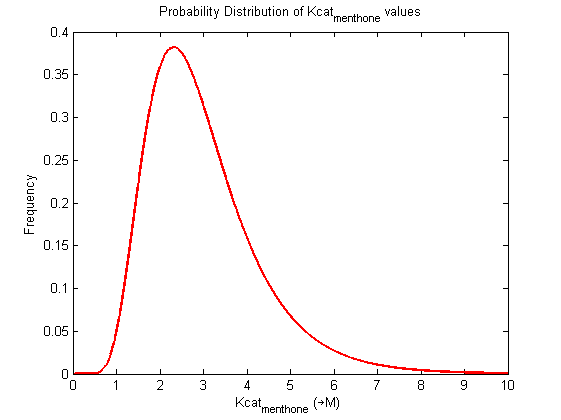
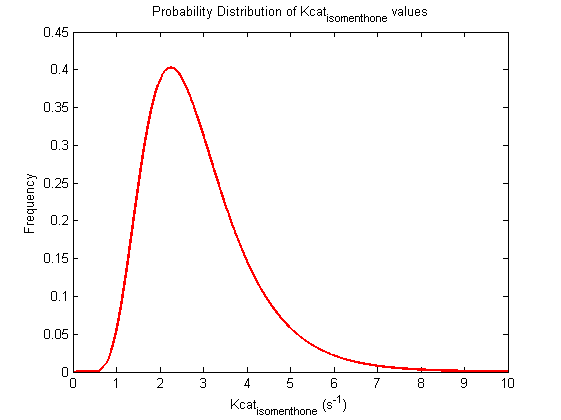
Parameter uncertainty
Probability distribution has been generated for each of the parameter in this reaction.
Detailed description of kinetic values obtained from literature
A more detailed description of the values listed above can be found here .
Simulations
References
- ↑ Davis, E.M. et. al. (2005). "Monoterpene metabolism. Cloning, Expression, and Characterization of Menthone Reductases from Peppermint.", Vol 137, pp. 873-881
- ↑ Kjonaas, R. et. al. (1982). Metabolism of monoterpenes: Conversion of l-Menthone to l-Menthol and d-Neomenthol by stereospecific dehydrogenases from peppermint (Mentha piperita) leaves, vol 69, pp.1013-1017.
- ↑ Latendresse M. (2013). "Computing Gibbs Free Energy of Compounds and Reactions in MetaCyc."
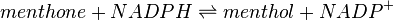
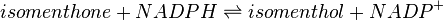
![V_\mathrm{MNMR} = Kcat_\mathrm{forward} * [MNMR] * \cfrac {\left ( \cfrac{[menthone]}{Km_\mathrm{menthone}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[neomenthol]*[NADP]}{[menthone]*[NADPH]*K_\mathrm{eq}} \right )}
{ \left (1 + \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[menthone]}{Km_\mathrm{menthone}} + \cfrac {[neomenthol]}{Km_\mathrm{neomenthol}} \right ) }](/wiki/images/math/4/e/5/4e5327f1b638bd8277017bae940a12d2.png)
![V_\mathrm{MNMR} = Kcat_\mathrm{forward} * [MNMR] * \cfrac {\left ( \cfrac{[isomenthone]}{Km_\mathrm{isomenthone}} * \cfrac {[NADPH]}{Km_\mathrm{NADPH}} \right ) * \left ( 1 - \cfrac {[neoisomenthol]*[NADP]}{[isomenthone]*[NADPH]*K_\mathrm{eq}} \right )}
{ \left (1 + \cfrac {[NADPH]}{Km_\mathrm{NADPH}} + \cfrac {[NADP]}{Km_\mathrm{NADP}} \right ) + \left ( 1+ \cfrac {[isomenthone]}{Km_\mathrm{isomenthone}} + \cfrac {[neoisomenthol]}{Km_\mathrm{neoisomenthol}} \right ) }](/wiki/images/math/7/2/2/722698a648d89f89e2d5b070258f0c7c.png)